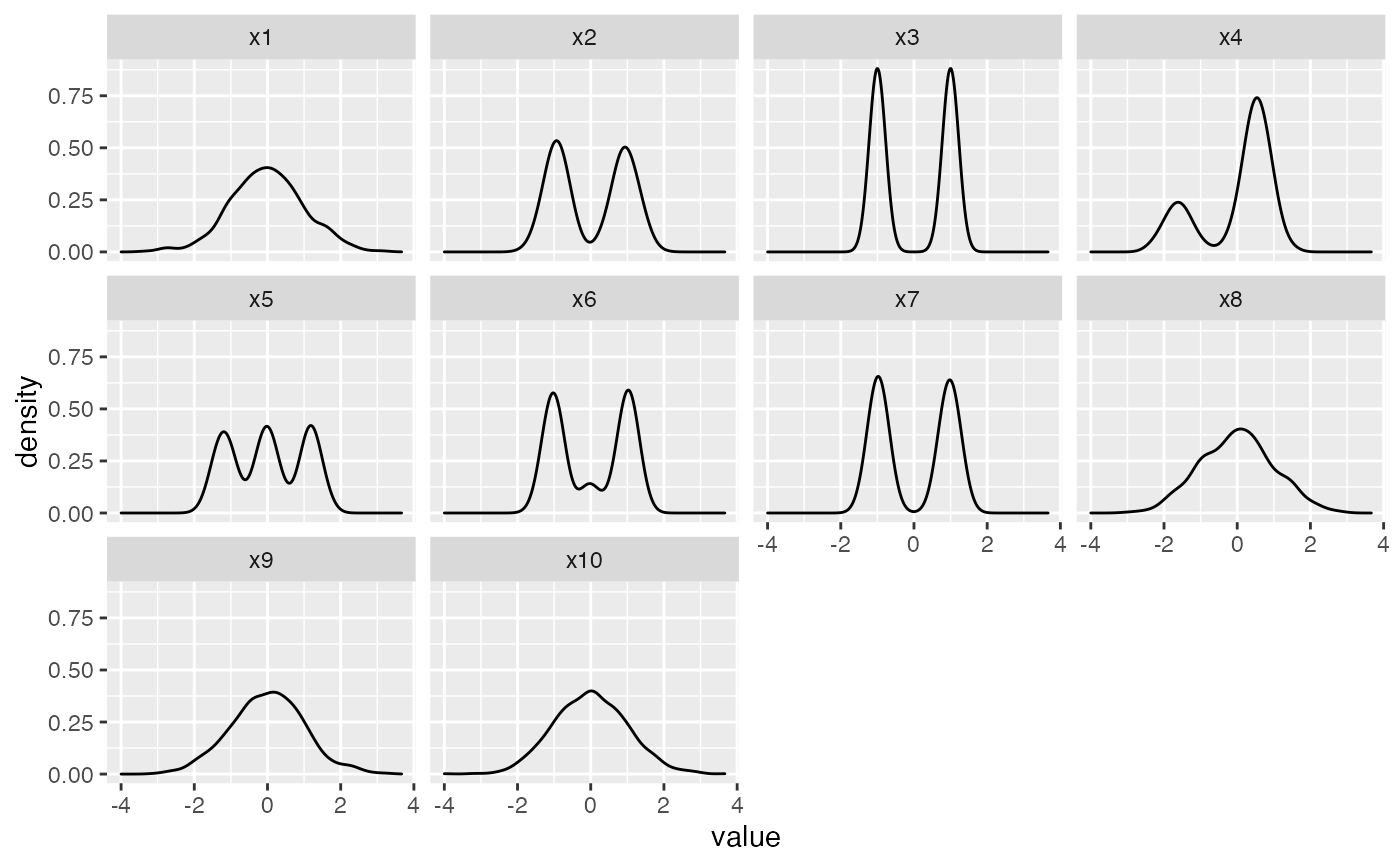
Simulated data to demonstrate the usage of four diagnostic plots in the package, users can create their own guided tour data objects and diagnose with the visualisation designed in this package.
Usage
holes_1d_geo
holes_1d_better
holes_1d_jellyfish
holes_2d_jellyfish
holes_2d_better
holes_2d_better_max_triesFormat
An object of class tbl_df (inherits from tbl, data.frame) with 416 rows and 8 columns.
An object of class tbl_df (inherits from tbl, data.frame) with 79 rows and 8 columns.
An object of class tbl_df (inherits from tbl, data.frame) with 2500 rows and 8 columns.
An object of class tbl_df (inherits from tbl, data.frame) with 2500 rows and 8 columns.
An object of class tbl_df (inherits from tbl, data.frame) with 98 rows and 8 columns.
An object of class tbl_df (inherits from tbl, data.frame) with 1499 rows and 8 columns.
Details
The prefix holes_* indicates the use of holes index in the guided tour.
The suffix *_better/geo/jellyfish indicates the optimiser used:
search_better, search_geodesic, search_jellyfish.
Examples
holes_1d_better %>%
explore_trace_interp(interp_size = 2) +
scale_color_continuous_botanical(palette = "fern")
#> map id to the x-axis
#> map tries to color