# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsElements of Data Science
SDS 322E
Department of Statistics and Data Sciences
The University of Texas at Austin
Fall 2025
Learning objectives
Understand the concept of tidy data: identify whether a dataset is tidy or not (we will cover how to tidy a messy data in code in week 4).
Understand the pipe operator (
|>): you need to know how to read and think about code with pipes.
Seek helps during the office hours
GDC (Gates Dell Complex) level 7 open space
- Monday 10-12pm after class with me
- Tuesday 2-3:15pm with Luke Bellinger (UGCA)
- Wednesday 3-5pm with Arka Sinha (Grad TA)
- Friday 10-11am with Luke Bellinger (UGCA)
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Tidy data

- Each variable is a column
- Each observation is a row
- Each cell is a single value
Is this tidy? (1/8)
income and religion in the US produced by Pew Research Center in 2014
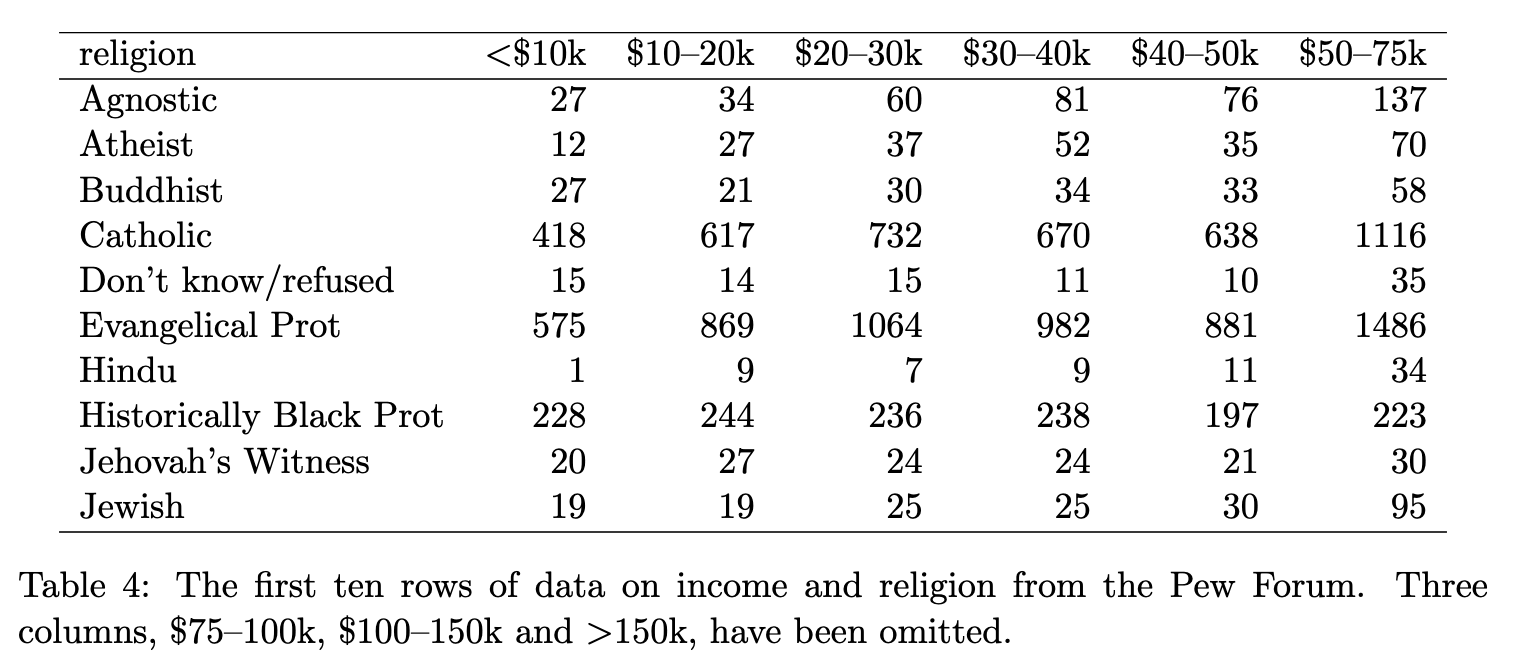
❌ No, because values (<$10k, $10-20k, $20-30k, …) are in variable names
Is this tidy? (2/8)
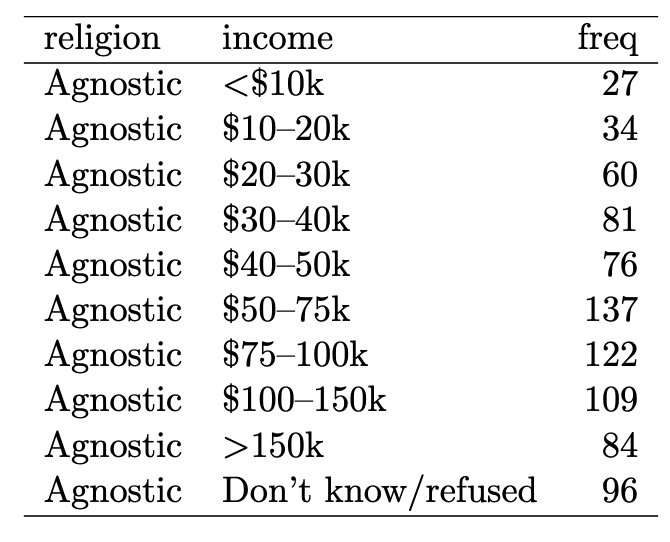
✅ Yes, because 1) The variables are: religion, income, and freq (count), 2) The observation is a demographic unit corresponding to a combination of religion and income
Is this tidy? (3/8)
The Billboard dataset: the date a song first entered the Billboard Top 100
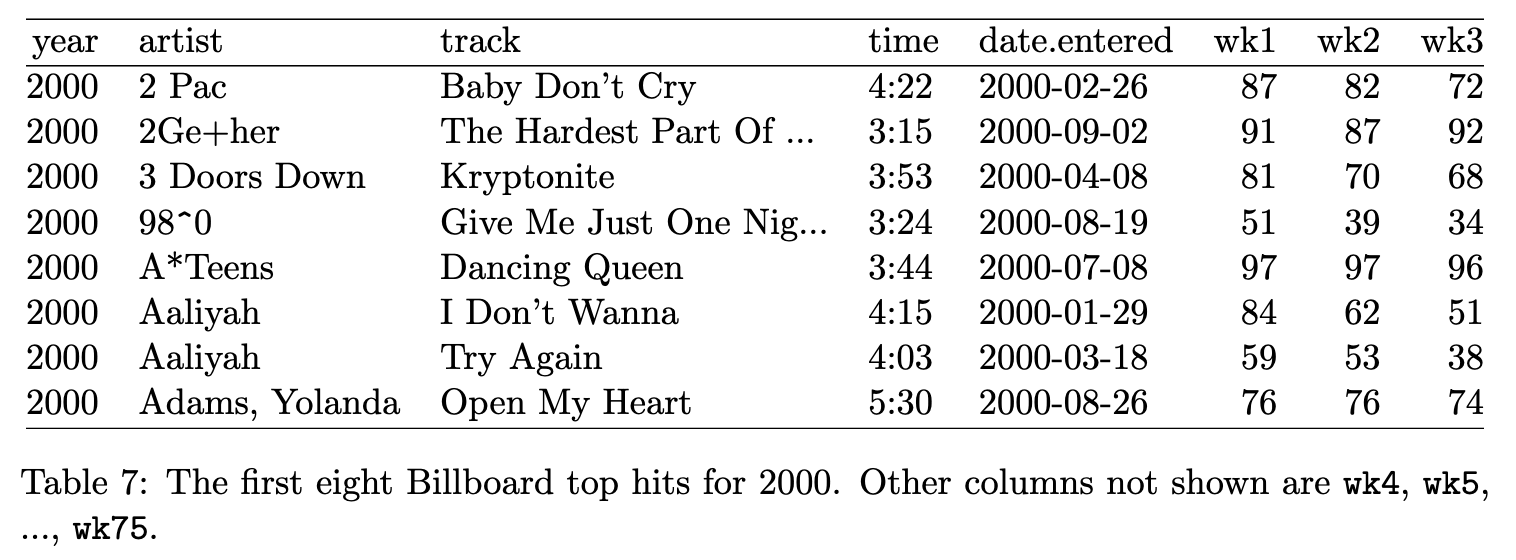
❌: No, because wk1, wk2, … are values, not variables - they should be recorded in cells rather than in column names
Is this tidy? (4/8)
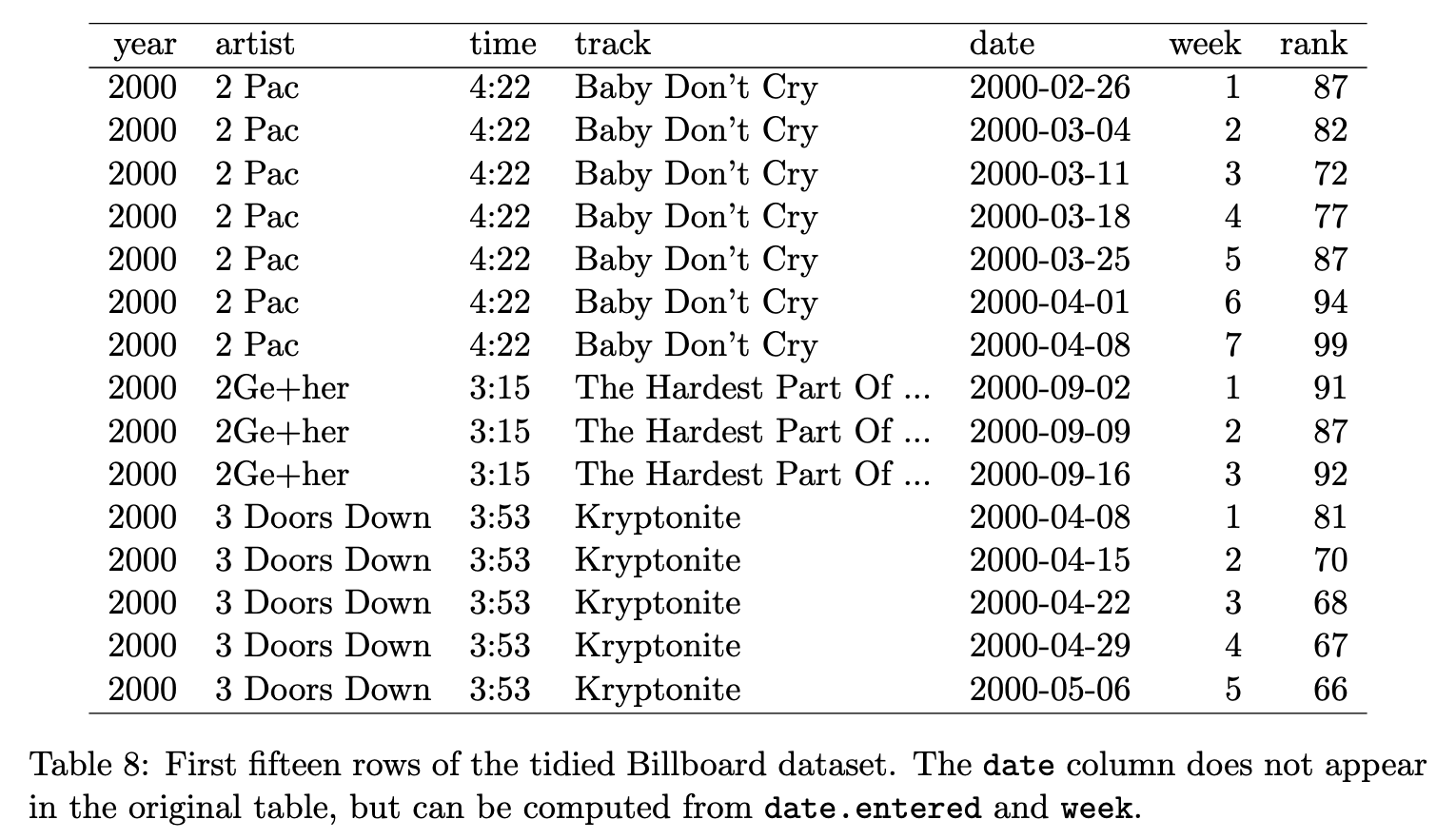
✅ Yes, because 1) The variables are: year, artist, time, track, date, week, and rank, 2) The observation is a recorded rank of a song in a particular week
Is this tidy? (5/8)
Number of cases of TB (tuberculosis)
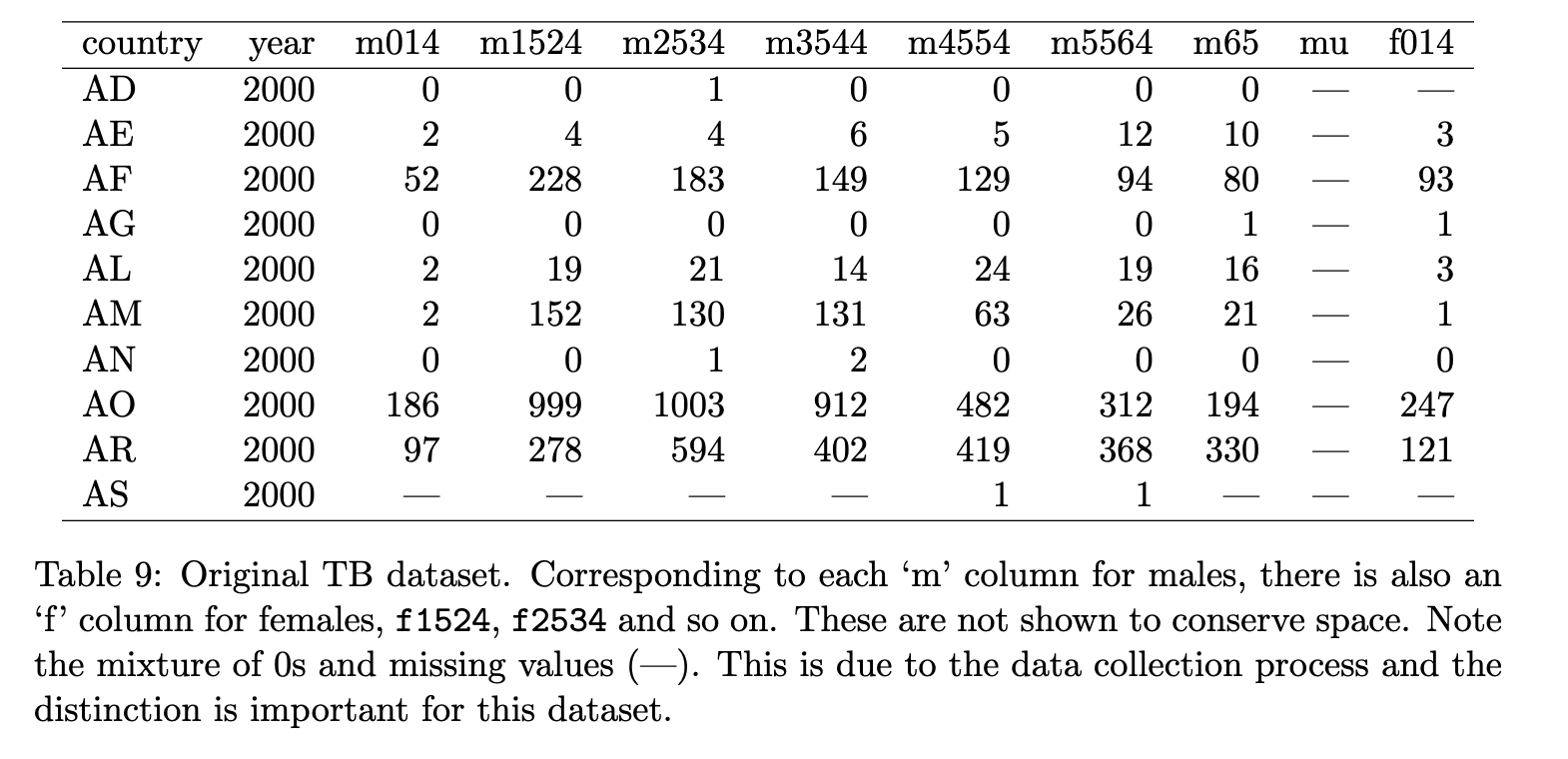
Some information: m014 means for male, 1-14 year old, m1524 means for male 15-24 year old, etc.
❌ No, because the column names contain multiple variable names: gender (m/f) and age (both lower end and higher end of the range).
Is this tidy? (6/8)
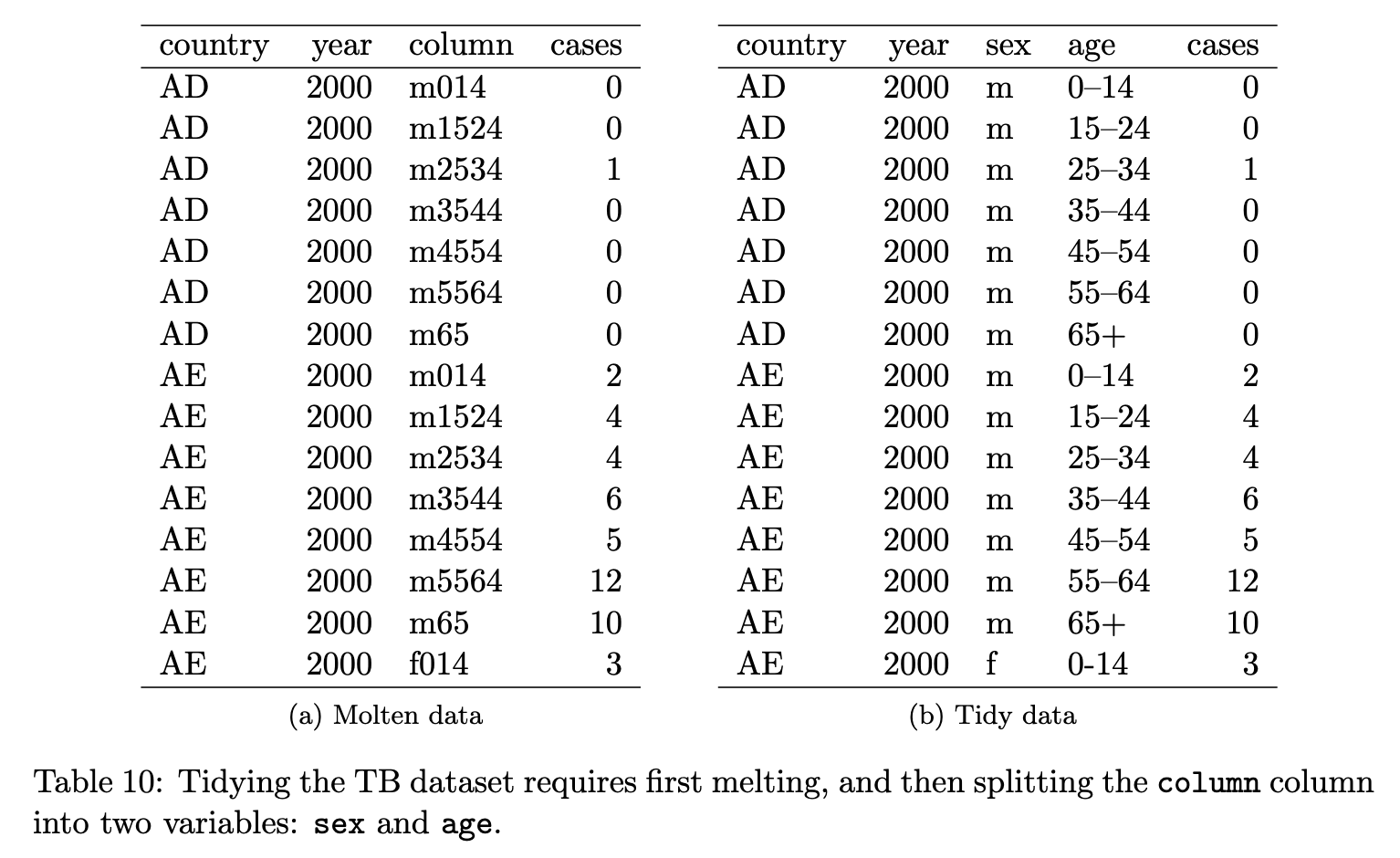
✅ Yes, because 1) The variables are: country, year, column, and cases, and 2) the observation is the number of cases per year, per gender age group, per country
Both are tidy data - you will learn in week 4 how to clean it from (a) to (b)
Is this tidy? (7/8) - the gapminder data
✅ Yes, because
Each variable forms a column: country, continent, year, lifeExp, pop, gdpPercap
Each observation forms a row: each row is a country in a particular year
Each value forms a cell: e.g. life expectancy of Afghanistan in 1952 is 28.801
Is this tidy? (8/8) - the flight data
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time
<int> <int> <int> <int> <int> <dbl> <int>
1 2013 1 1 517 515 2 830
2 2013 1 1 533 529 4 850
3 2013 1 1 542 540 2 923
4 2013 1 1 544 545 -1 1004
5 2013 1 1 554 600 -6 812
6 2013 1 1 554 558 -4 740
7 2013 1 1 555 600 -5 913
8 2013 1 1 557 600 -3 709
9 2013 1 1 557 600 -3 838
10 2013 1 1 558 600 -2 753
# ℹ 336,766 more rows
# ℹ 12 more variables: sched_arr_time <int>, arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>,
# dest <chr>, air_time <dbl>, distance <dbl>, hour <dbl>,
# minute <dbl>, time_hour <dttm>✅ Yes, because 1) each variable forms a column, 2) each observation forms a row: each row is one flight, and 3) each value forms a cell
The pipe operator
If you download the Friday code, you will see something like this:
gapminder$country |> unique() |> length()
Once upon a time
| Abstraction | Example |
|---|---|
FUN_1(DATA) |
unique(gapminder$lifeExp) |
FUN_2(FUN_1(DATA)) |
length(unique(gapminder$lifeExp)) |
Abstraction: FUN_1(DATA, arg1 = val1, arg2 = val2)
Example: mutate(mtcars, kpl = mpg * 0.425)
Abstraction: FUN_2(FUN_1(DATA, arg1 = val1, arg2 = val2), arg3 = val3)
Example: filter(mutate(mtcars, kpl = mpg * 0.425), vs ==0)
Abstraction: FUN_3(FUN_2(FUN_1(DATA, arg1 = val1, arg2 = val2), arg3 = val3), arg4 = val4)
Example: group_by(filter(mutate(mtcars, kpl = mpg * 0.425), vs ==0), cyl)
We could keep going on to make the code annoyingly long . . .
Abstraction: FUN_4(FUN_3(FUN_2(FUN_1(DATA, arg1 = val1, arg2 = val2), arg3 = val3), arg4 = val4), arg5 = val5)
Example: summarize(group_by(filter(mutate(mtcars, kpl = mpg * 0.425), vs ==0), cyl), disp = mean(disp, na.rm = TRUE), kpl = mean(kpl, na.rm = TRUE))|
With line breaks we can do:
We need to read from middle out 😿
Why this works?
The pipe operator abstracts out the first argument of a function, so
unique(gapminder$country)
gapminder$country |> unique()
This is a powerful abstraction that allows us to chain together a sequence of data transformations (aka dplyr functions) in a clear and readable way since all the tidyverse functions take the data frame as the first argument.
mtcarsis a data framemutate()is a function that takes the dataset as its first argument, so we can domtcars |> mutate(kpl = mpg * 0.425144)
- The output of above is still a data frame, so we can pipe it to the next function
filter():
mtcars |> mutate(kpl = mpg * 0.425144) |> filter(vs == 0)
Why this works?
- The output of this is still a data frame, so we can pipe it to the next function
group_by():
Pipe logistics
On your keyboard, the pipe operator is produced by:
|(vertical bar):shift+\(backslash), plus>(greater than):shift+.(period).
There is a shortcut: control + shift + M (for Mac: command + shift + M)
You will see a lot of pipes in the Friday class - be prepared!