# A tibble: 3 × 2
x y
<dbl> <chr>
1 1 a
2 2 b
3 3 c # A tibble: 3 × 2
x z
<dbl> <chr>
1 1 d
2 2 e
3 4 f left_join(), right_join(), inner_join(), full_join(), semi_join(), and anti_join()filter(), mutate(), select(), %in%, fct_reorder()tibble(), bind_rows(), fct_recode(), fct_relevel()Recall how the pipe operator |> works (from the Wednesday class in week 2):
Reminder:
You’re actually doing a very good job. The script for each class contains over 100 lines of code!
At the same time, if you find the exercise in the class overwhelming, come to the office hours!
How would you write filter(flights, month == 1) using the pipe operator?
flights |> filter(month == 1)
The first arguemnt of filter() is the dataset. flight is the dataset. We cna put it in the front of the function using the pipe operator.
What if you want to select the variable month, day, and year from the filtered dataset?
flights |> filter(month == 1) |> select(month, day, year)
The output from flights |> filter(month == 1) is a tibble, which is the first argument for select(). We can put it in the front of the function.
Think of it as we take the flight data, filter it by month == 1, and then select the variables month, day, and year from the filtered data.
Mutate join
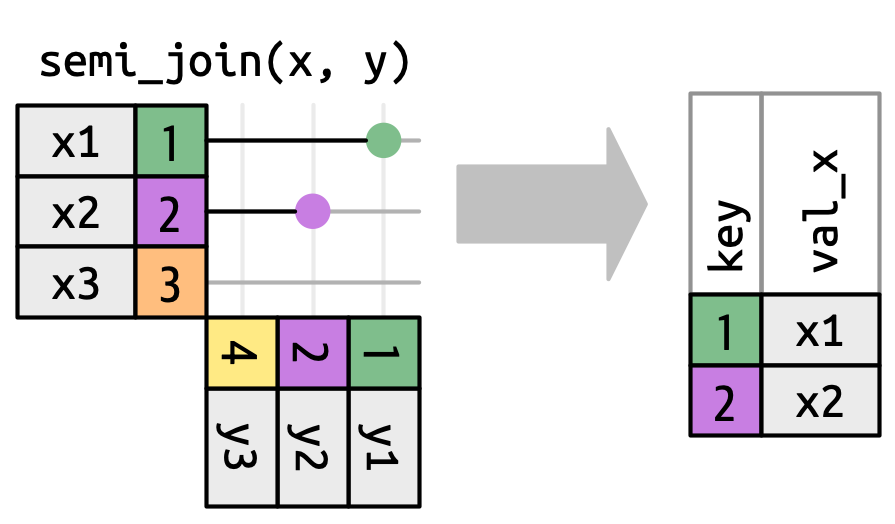
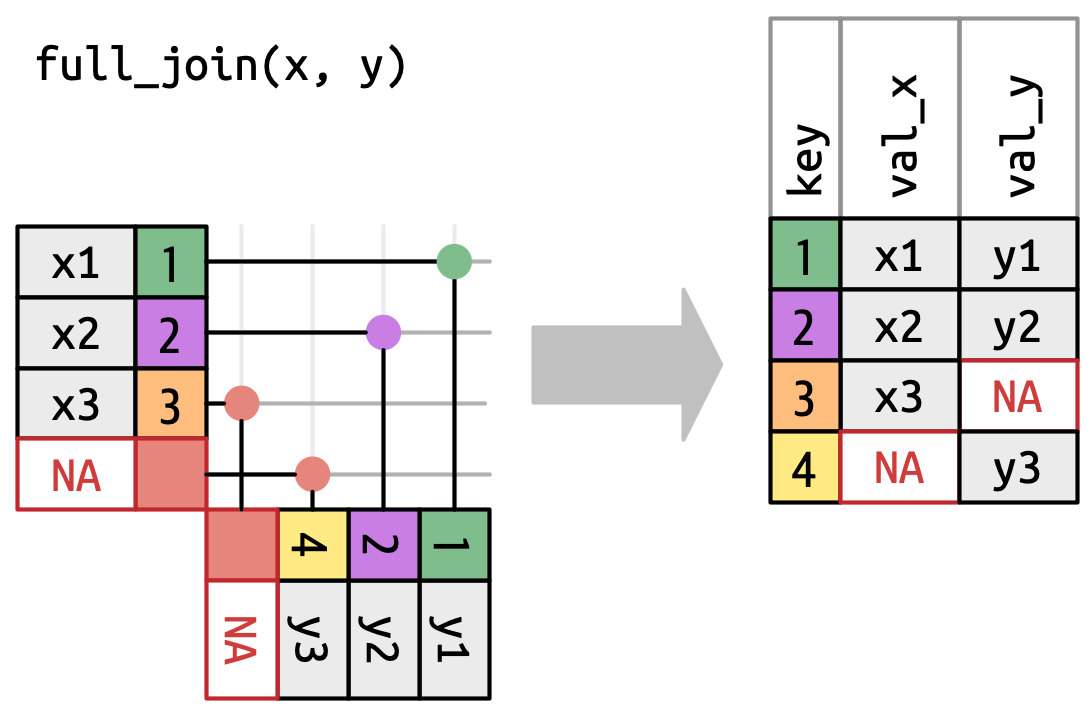
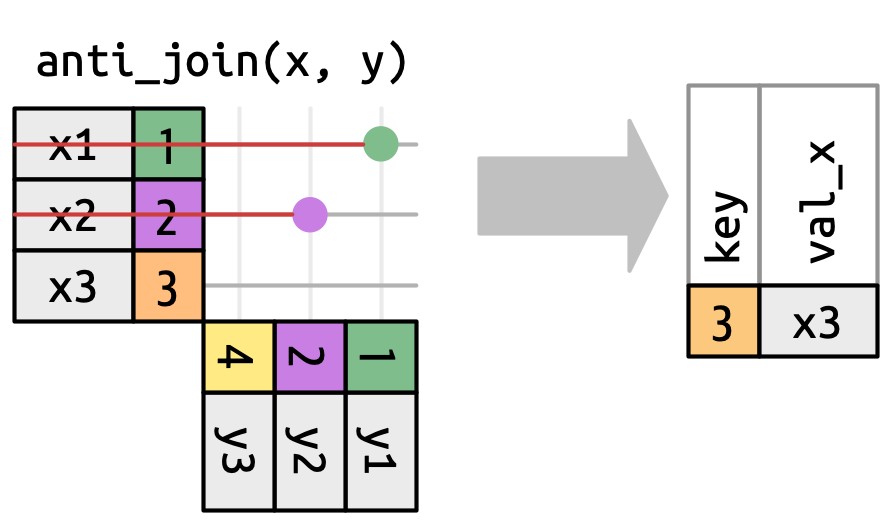
left_join()right_join()inner_join()full_join()Filter join
semi_join()anti_join()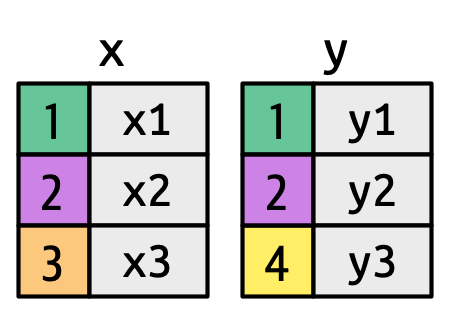
A left join keeps all observations in data frame x. Every row of x is preserved in the output and NA is used if there is no matching value in data frame y.
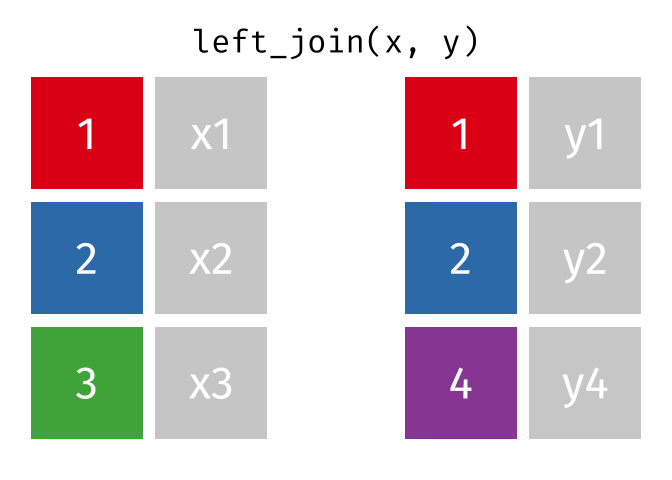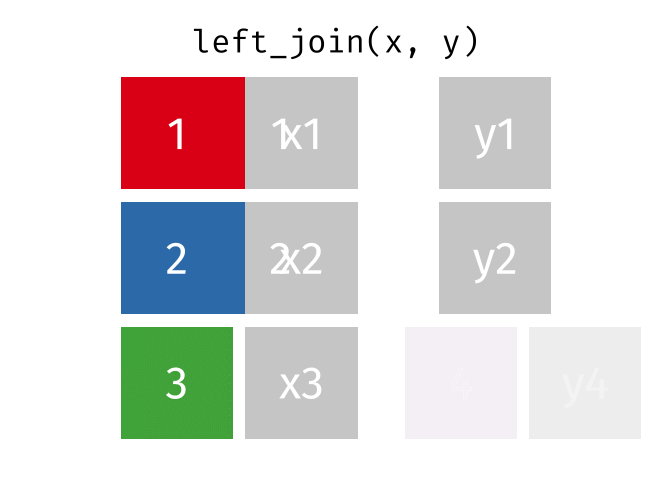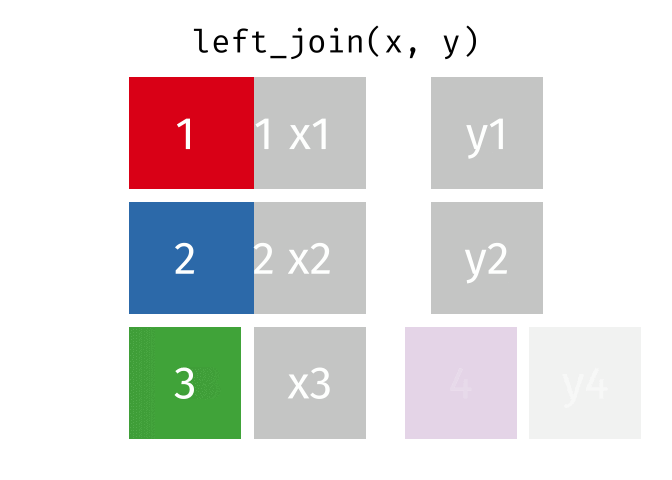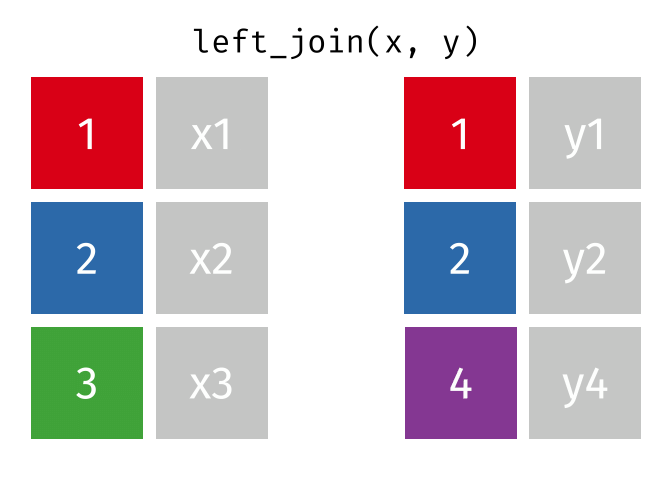
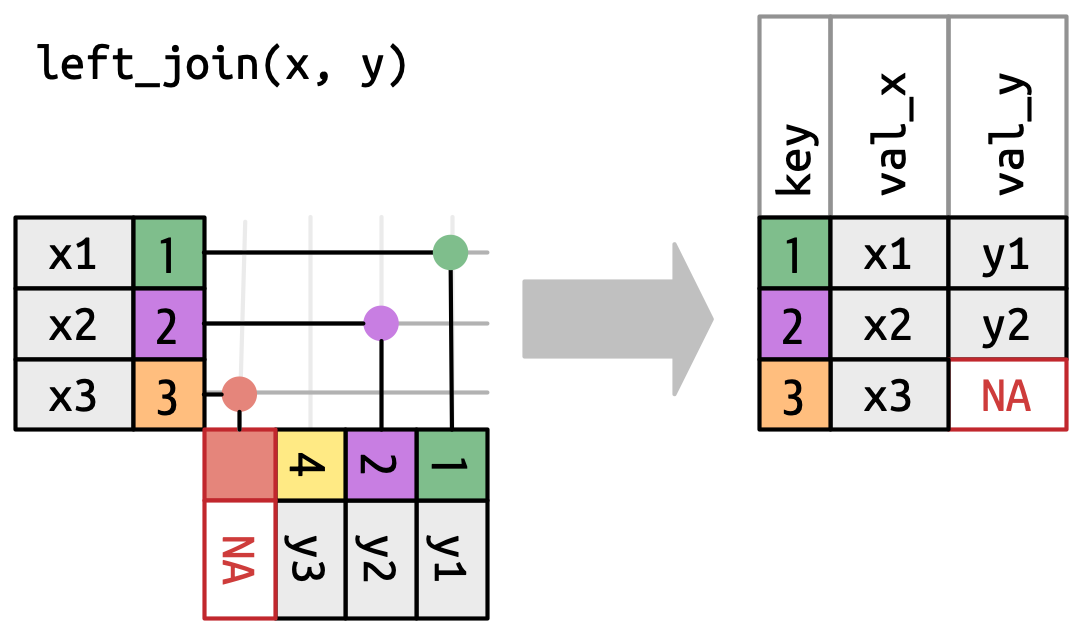
left_join(DATA_X, DATA_Y, by = "SHARED_KEY")
Other variations:
When the key variables have different names in the two data frames, you need to specify the names of the key variables in both data frames.
left_join(DATA_X, DATA_Y, by = c("KEY_IN_X" = "KEY_IN_Y"))
# A tibble: 3 × 3
x1 y z
<dbl> <chr> <chr>
1 1 a d
2 2 b e
3 3 c <NA> Other variations:
Auto-detect won’t work if the key variables have different names.
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
# ℹ 336,771 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>flights_tiny <- flights |>
select(month:day, hour, origin,
dest, tailnum, carrier) |>
head(5)
flights_tiny# A tibble: 5 × 7
month day hour origin dest tailnum carrier
<int> <int> <dbl> <chr> <chr> <chr> <chr>
1 1 1 5 EWR IAH N14228 UA
2 1 1 5 LGA IAH N24211 UA
3 1 1 5 JFK MIA N619AA AA
4 1 1 5 JFK BQN N804JB B6
5 1 1 6 LGA ATL N668DN DL flights_sml <- flights |>
select(month:day, hour, origin,
dest, tailnum, carrier) |>
head(30)
flights_sml# A tibble: 30 × 7
month day hour origin dest tailnum carrier
<int> <int> <dbl> <chr> <chr> <chr> <chr>
1 1 1 5 EWR IAH N14228 UA
2 1 1 5 LGA IAH N24211 UA
3 1 1 5 JFK MIA N619AA AA
4 1 1 5 JFK BQN N804JB B6
5 1 1 6 LGA ATL N668DN DL
# ℹ 25 more rows# A tibble: 1,458 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 04G Lansd… 41.1 -80.6 1044 -5 A Amer…
2 06A Moton… 32.5 -85.7 264 -6 A Amer…
3 06C Schau… 42.0 -88.1 801 -6 A Amer…
4 06N Randa… 41.4 -74.4 523 -5 A Amer…
5 09J Jekyl… 31.1 -81.4 11 -5 A Amer…
# ℹ 1,453 more rows# A tibble: 3 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 EWR Newar… 40.7 -74.2 18 -5 A Amer…
2 JFK John … 40.6 -73.8 13 -5 A Amer…
3 LGA La Gu… 40.8 -73.9 22 -5 A Amer…origin variable in the flights_tiny data is connected to the faa variable in the airports_tiny data.# A tibble: 5 × 14
month day hour origin dest tailnum carrier name lat lon alt tz
<int> <int> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 1 5 EWR IAH N14228 UA Newark… 40.7 -74.2 18 -5
2 1 1 5 LGA IAH N24211 UA La Gua… 40.8 -73.9 22 -5
3 1 1 5 JFK MIA N619AA AA John F… 40.6 -73.8 13 -5
4 1 1 5 JFK BQN N804JB B6 John F… 40.6 -73.8 13 -5
5 1 1 6 LGA ATL N668DN DL La Gua… 40.8 -73.9 22 -5
# ℹ 2 more variables: dst <chr>, tzone <chr>Still the same two data, now I want to join by the destination airport.
# A tibble: 5 × 14
month day hour origin dest tailnum carrier name lat lon alt tz
<int> <int> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 1 5 EWR IAH N14228 UA George… 30.0 -95.3 97 -6
2 1 1 5 LGA IAH N24211 UA George… 30.0 -95.3 97 -6
3 1 1 5 JFK MIA N619AA AA Miami … 25.8 -80.3 8 -5
4 1 1 5 JFK BQN N804JB B6 <NA> NA NA NA NA
5 1 1 6 LGA ATL N668DN DL Hartsf… 33.6 -84.4 1026 -5
# ℹ 2 more variables: dst <chr>, tzone <chr>It seems that we don’t have BQN matched from the airports_sml data.
BQN is the IATA code for Rafael Hernández Airport, located in Aguadilla, Puerto Rico.
Option 1: Let’s only focus on the flights in the continental US.
# A tibble: 4 × 14
month day hour origin dest tailnum carrier name lat lon alt tz
<int> <int> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 1 5 EWR IAH N14228 UA George… 30.0 -95.3 97 -6
2 1 1 5 LGA IAH N24211 UA George… 30.0 -95.3 97 -6
3 1 1 5 JFK MIA N619AA AA Miami … 25.8 -80.3 8 -5
4 1 1 6 LGA ATL N668DN DL Hartsf… 33.6 -84.4 1026 -5
# ℹ 2 more variables: dst <chr>, tzone <chr>Let’s add the Puerto Rico airport information to the airports data.
bqn_tbl <- tibble(faa = "BQN", name = "Rafael Hernández Airport",
lat = 18.4944, lon = -67.1296, alt = 49, tz = -4,
dst = "A", tzone = "America/Puerto_Rico")
bqn_tbl# A tibble: 1 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 BQN Rafael Hernández Airport 18.5 -67.1 49 -4 A America/Puerto_R…# A tibble: 3 × 8
faa name lat lon alt tz dst tzone
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 ZWU Washington Union Station 38.9 -77.0 76 -5 A America/New_York
2 ZYP Penn Station 40.8 -74.0 35 -5 A America/New_York
3 BQN Rafael Hernández Airport 18.5 -67.1 49 -4 A America/Puerto_R…# A tibble: 5 × 14
month day hour origin dest tailnum carrier name lat lon alt tz
<int> <int> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 1 5 EWR IAH N14228 UA George… 30.0 -95.3 97 -6
2 1 1 5 LGA IAH N24211 UA George… 30.0 -95.3 97 -6
3 1 1 5 JFK MIA N619AA AA Miami … 25.8 -80.3 8 -5
4 1 1 5 JFK BQN N804JB B6 Rafael… 18.5 -67.1 49 -4
5 1 1 6 LGA ATL N668DN DL Hartsf… 33.6 -84.4 1026 -5
# ℹ 2 more variables: dst <chr>, tzone <chr>Without running the code, guess whether the following code will work.
Part 1
select() is a function from the dplyr package that keeps (drops) columns from a data frame.
Part 2
unique() is a function from the base package that find the unique value in a vector.
In week 1 lecture 3, we use it to check the number of unique continent in the gapminder data (page 19/32).
Grab the class repository:
Connect the flights_sml data with the airport information by the destination airport.
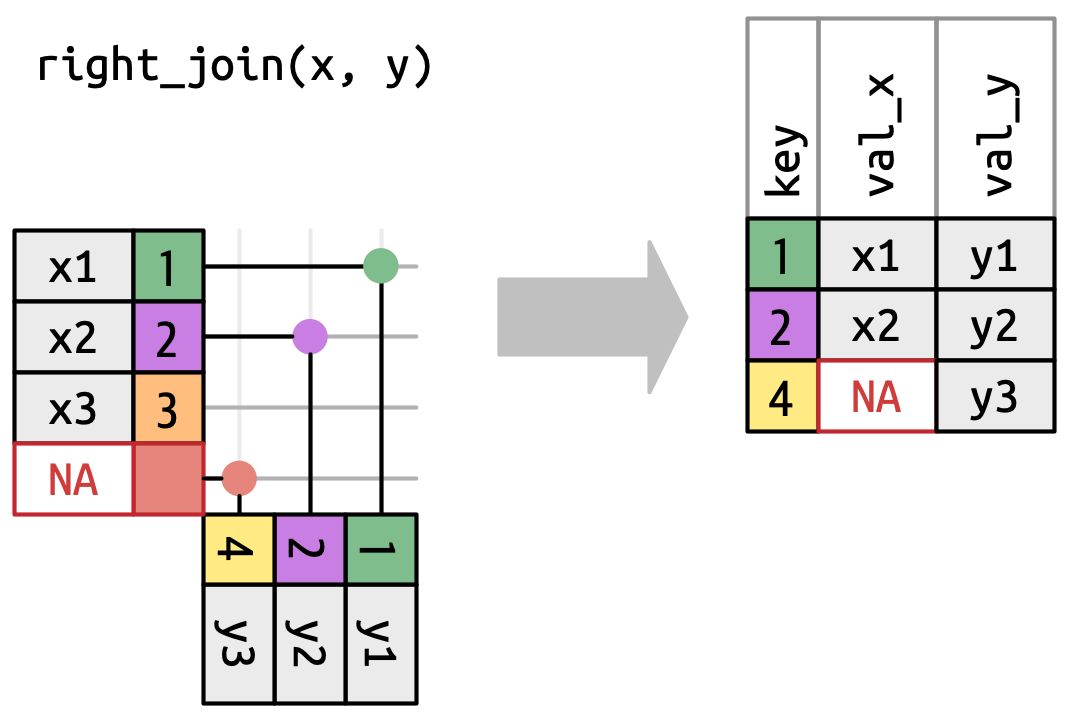
A left join keeps all observations in x. Every row of x is preserved in the output and NA is used if there is no matching value in y.
An inner join retained the rows if and only if the keys are equal.
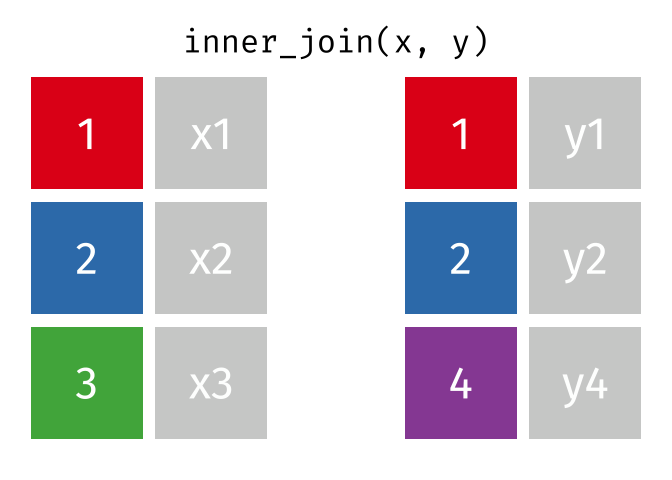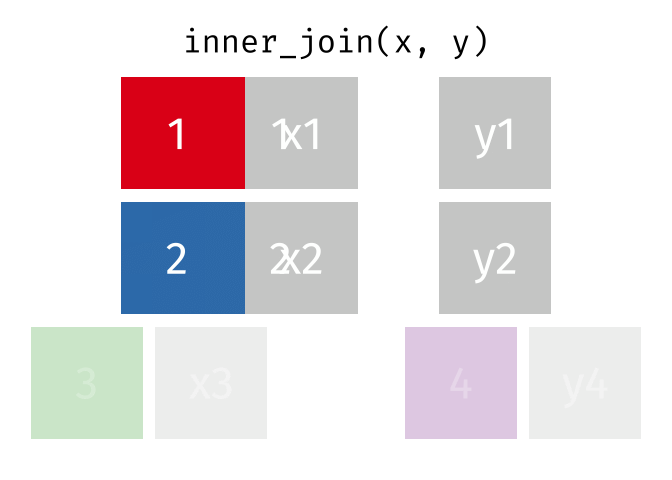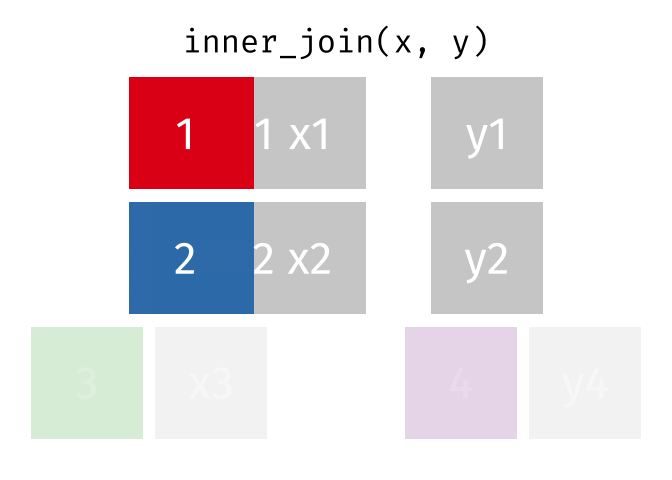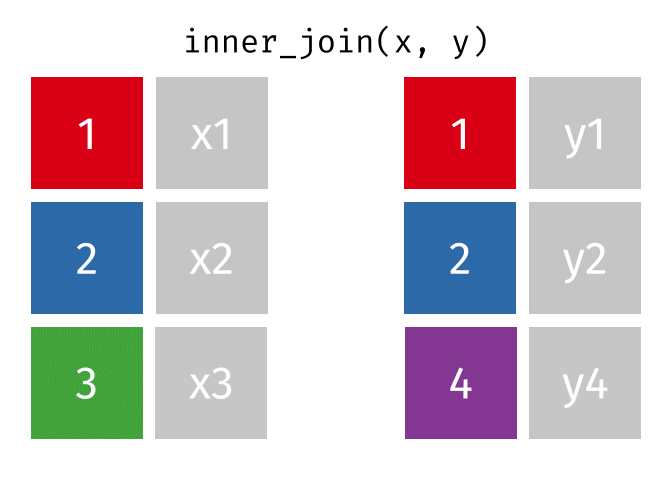
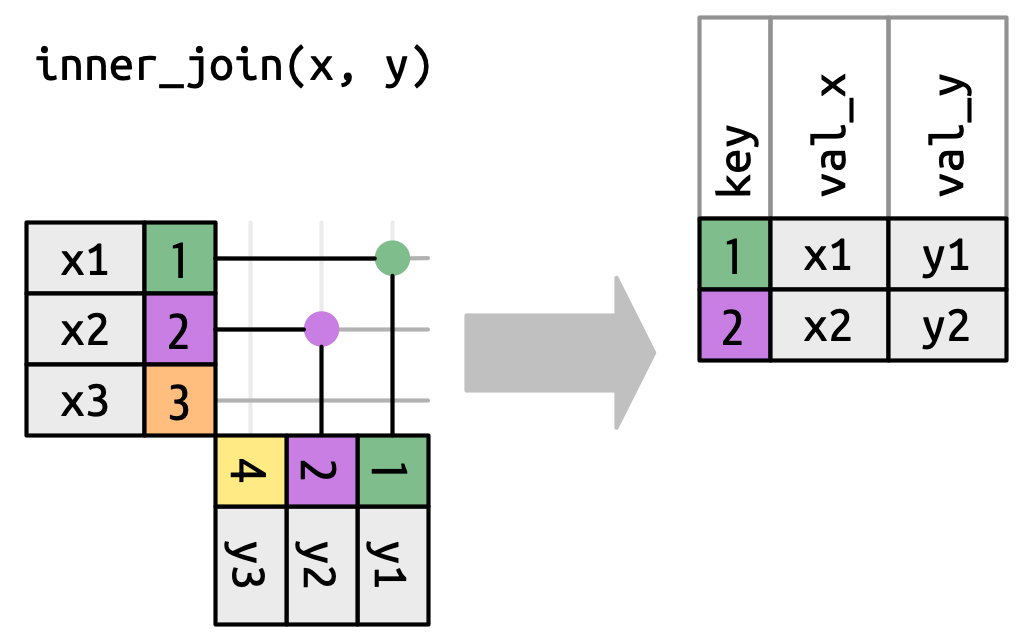
What would you expect the result to be if we do an inner join rather than left join?
# A tibble: 4 × 14
month day hour origin dest tailnum carrier name lat lon alt tz
<int> <int> <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 1 1 5 EWR IAH N14228 UA George… 30.0 -95.3 97 -6
2 1 1 5 LGA IAH N24211 UA George… 30.0 -95.3 97 -6
3 1 1 5 JFK MIA N619AA AA Miami … 25.8 -80.3 8 -5
4 1 1 6 LGA ATL N668DN DL Hartsf… 33.6 -84.4 1026 -5
# ℹ 2 more variables: dst <chr>, tzone <chr>Only the four rows that have a match in the airports_dest data are kept.
flights data?Maybe I want a better facet header than “AA”, “DL”, and “UA”. 🤔
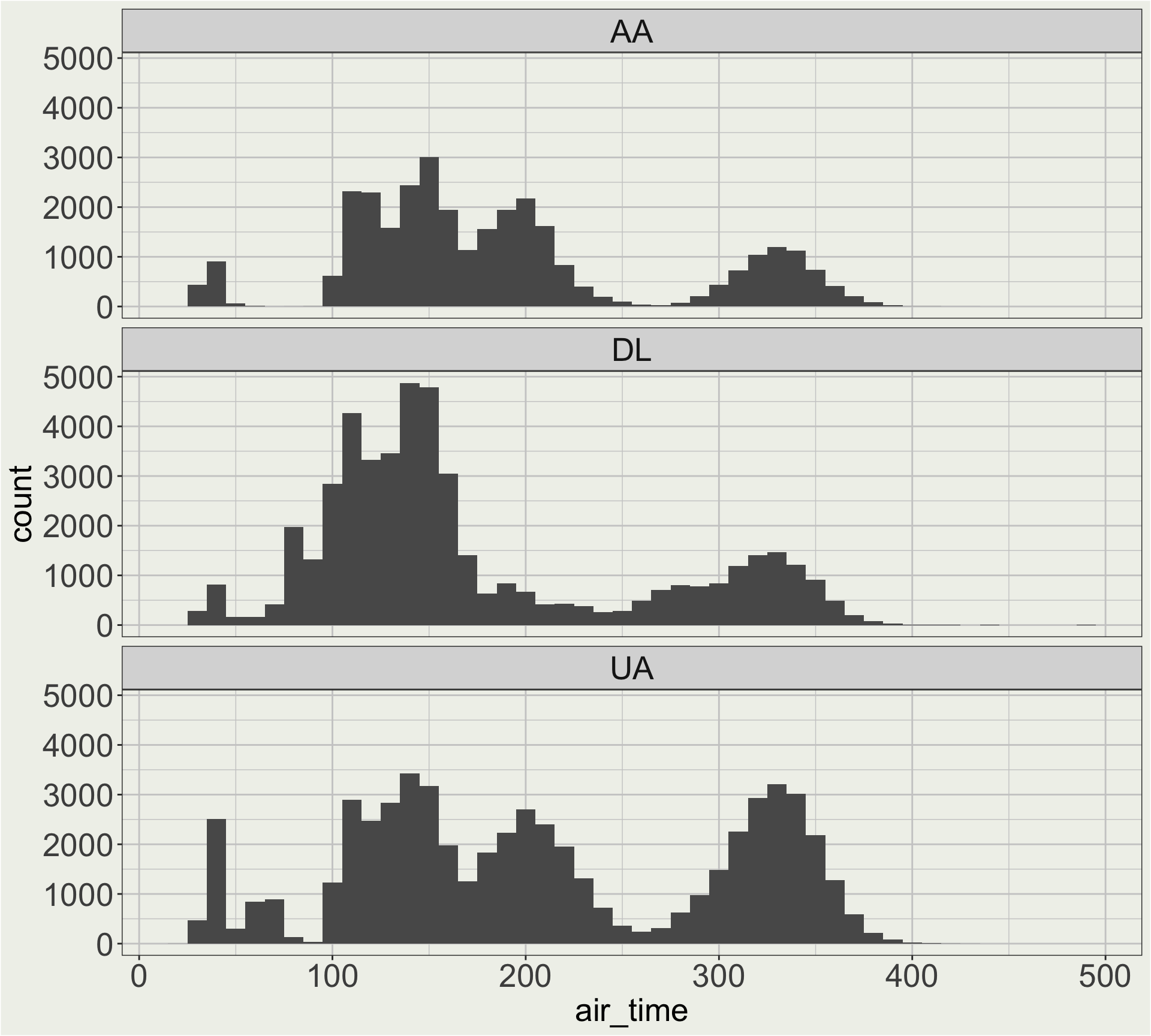
I didn’t give you can example on fct_recode() but here you go.
flights |>
# we looked in the air_time vs. distance plot
# before the there are long flights
# thqt go to HNL (Hawaii)
filter(dest != "HNL") |>
filter(carrier %in% c("AA", "DL", "UA")) |>
mutate(carrier = fct_recode(
carrier,
"American Airlines" = "AA",
"Delta Air Lines" = "DL",
"United Air Lines" = "UA")) |>
ggplot(aes(x = air_time)) +
geom_histogram(binwidth = 10) +
facet_wrap(vars(carrier), ncol = 1)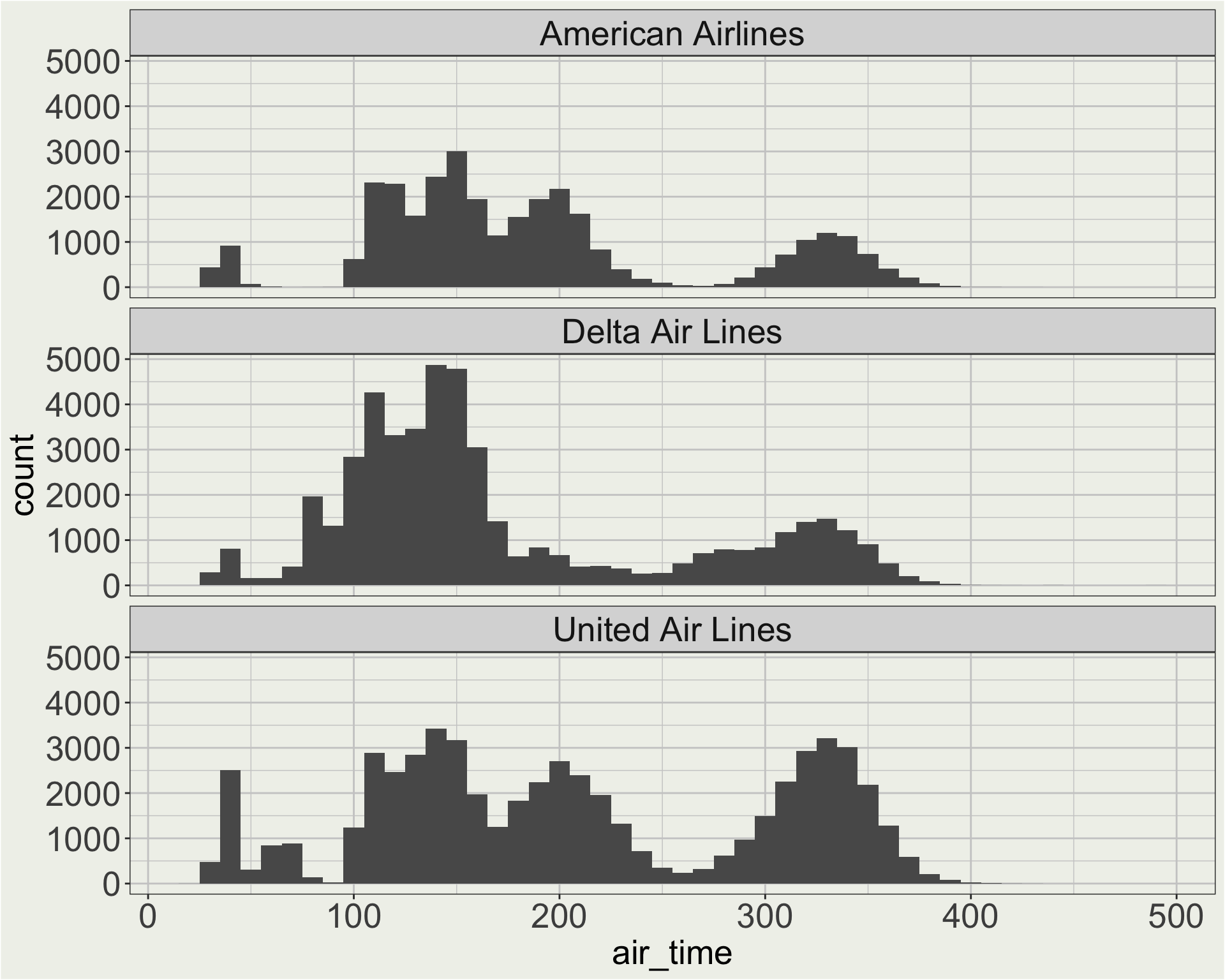
Often, we make a base plot first (previous page), see somewhere we want to improve (facet header), and then go back to the data wrangling step to make the change (mutate(carrier = fct_recode(...))).
When we have more carriers, manually recode all the airlines becomes tedious. 🤔
# A tibble: 3 × 2
carrier name
<chr> <chr>
1 9E Endeavor Air Inc.
2 AA American Airlines Inc.
3 AS Alaska Airlines Inc. our_carriers = c("AA", "DL", "WN",
"MQ", "UA", "9E")
df <- flights |>
filter(dest != "HNL") |>
filter(carrier %in% our_carriers) |>
left_join(airlines, by = c("carrier" = "carrier"))
colnames(df) [1] "year" "month" "day"
[4] "dep_time" "sched_dep_time" "dep_delay"
[7] "arr_time" "sched_arr_time" "arr_delay"
[10] "carrier" "flight" "tailnum"
[13] "origin" "dest" "air_time"
[16] "distance" "hour" "minute"
[19] "time_hour" "name" 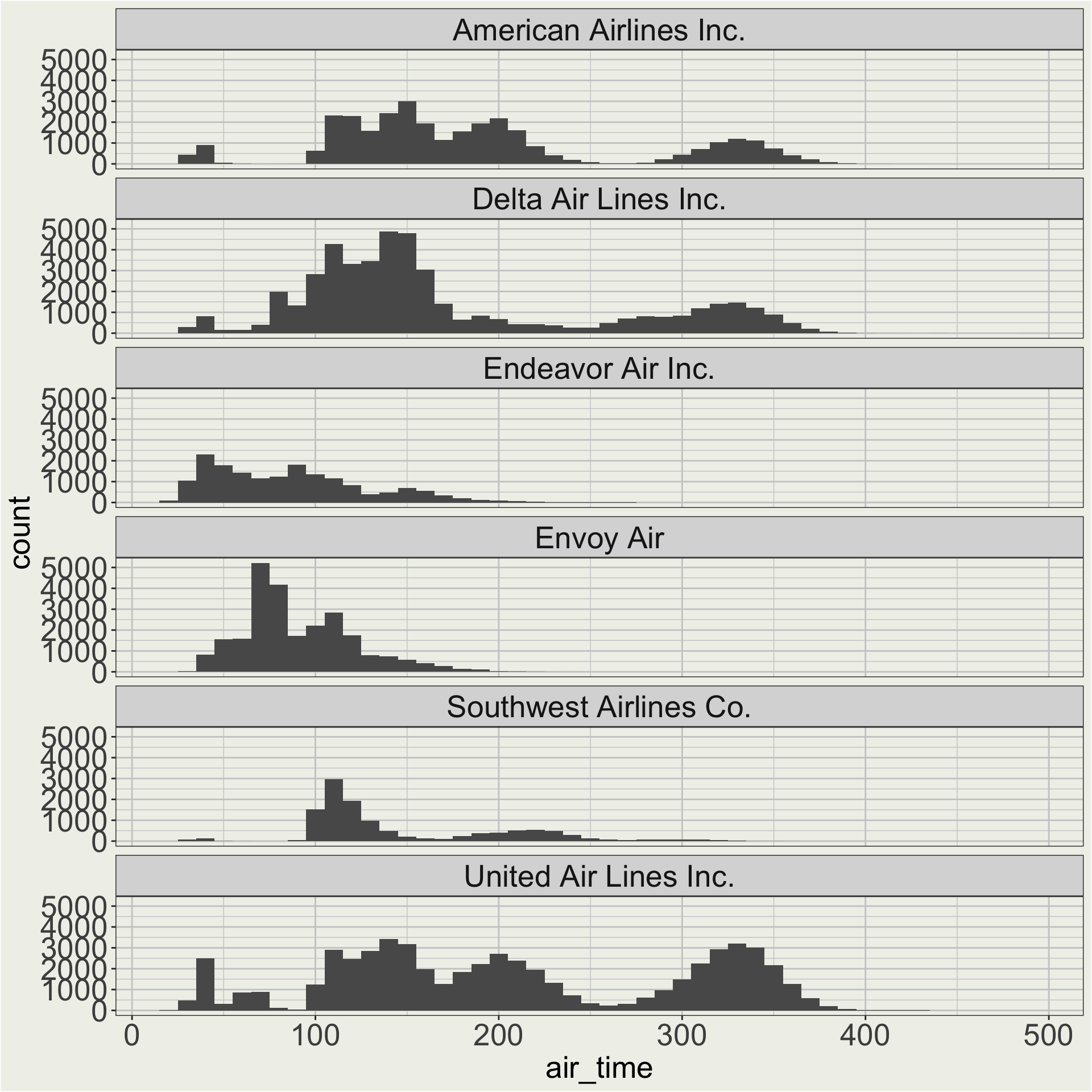
Here we care about the shape of the distribution of air time for each carrier, so maybe a free y scale is better.
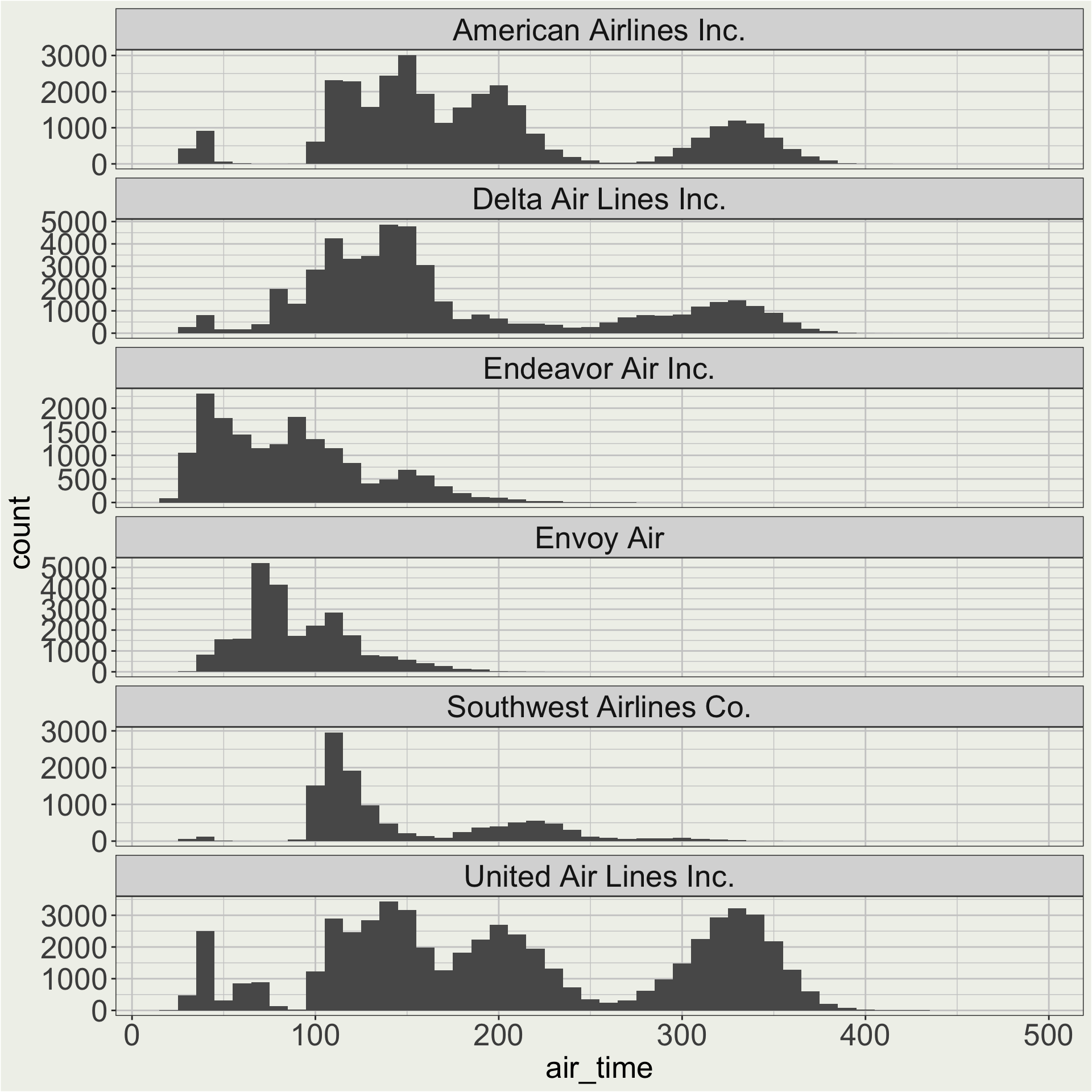
Maybe I want to change the order of the facet so that AA, DL, UA are on the top. 🤔
Technically, you can use fct_relevel() to name each factor level in the order you want.
We can also use fct_reorder() to order the factor levels by a numeric variable.
our_carriers = c("AA", "DL", "WN", "MQ", "UA", "9E")
df <- flights |>
filter(dest != "HNL") |>
filter(carrier %in% our_carriers) |>
left_join(airlines, by = c("carrier" = "carrier")) |>
mutate(name = fct_reorder(name, -air_time))
df |>
ggplot(aes(x = air_time)) +
geom_histogram(binwidth = 10) +
facet_wrap(vars(name), ncol = 1, scales = "free_y")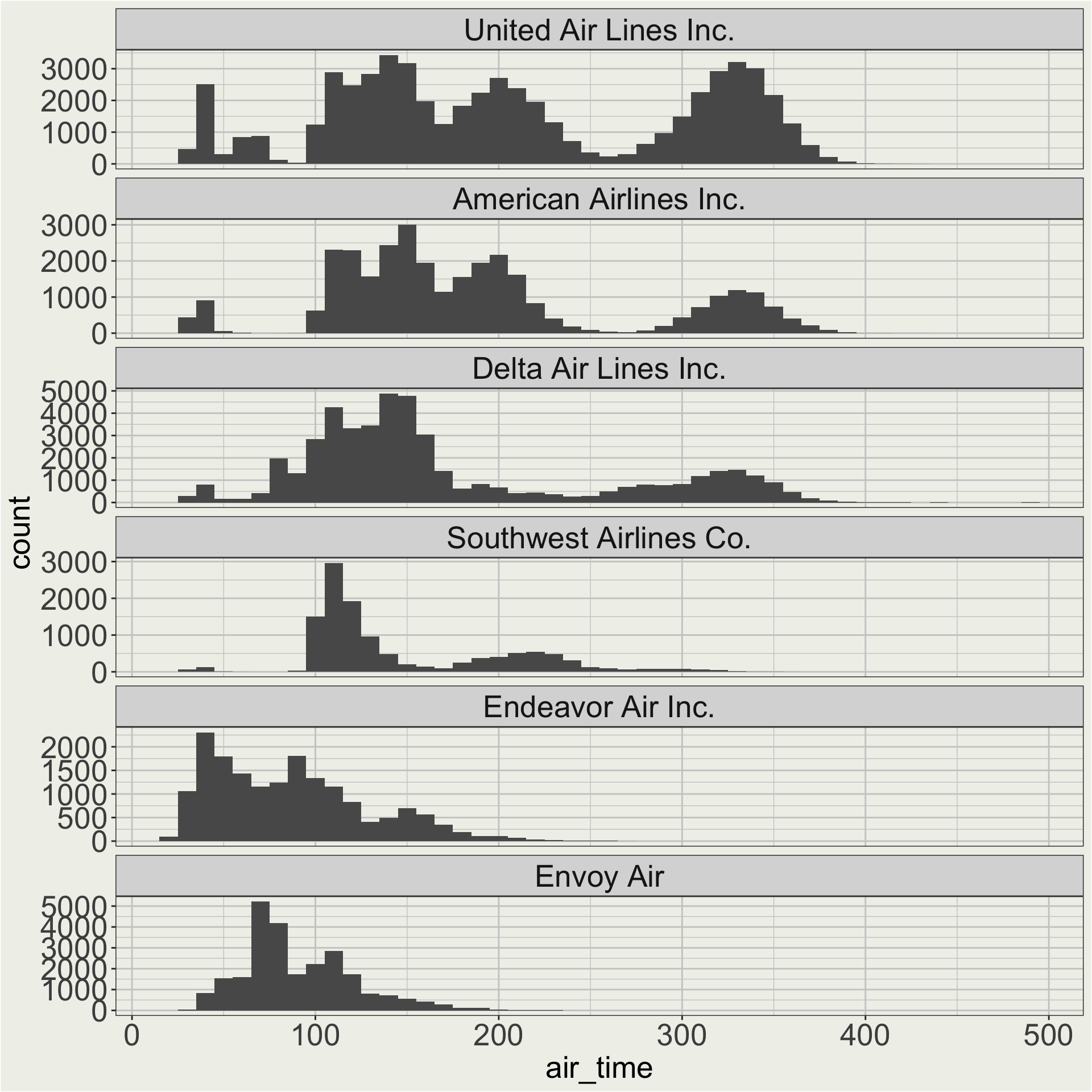
😃