# A tibble: 344 × 8
species island bill_len bill_dep flipper_len body_mass sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
2 Adelie Torgersen 39.5 17.4 186 3800 female 2007
3 Adelie Torgersen 40.3 18 195 3250 female 2007
4 Adelie Torgersen NA NA NA NA <NA> 2007
5 Adelie Torgersen 36.7 19.3 193 3450 female 2007
6 Adelie Torgersen 39.3 20.6 190 3650 male 2007
7 Adelie Torgersen 38.9 17.8 181 3625 female 2007
8 Adelie Torgersen 39.2 19.6 195 4675 male 2007
9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007
10 Adelie Torgersen 42 20.2 190 4250 <NA> 2007
# ℹ 334 more rowsElements of Data Science
SDS 322E
H. Sherry Zhang
Department of Statistics and Data Sciences
The University of Texas at Austin
Fall 2025
Department of Statistics and Data Sciences
The University of Texas at Austin
Fall 2025
Learning objectives
- Understand the hierarchical clustering algorithm
- Understand different linkage methods
- Apply hierarchical clustering to the data and compare the result with kmeans clustering
- calculate distance with
dist() - compute hierarchical clustering with
hclust() - plot the dendrogram:
ggdendrogram()(or base Rplot()) - cut the dendrogram with
cutree()
- calculate distance with
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Artwork by @allison_horst
Hierarchical clustering with the penguins data
Follow along the class example:
usethis::create_from_github(""SDS322E-2025Fall/0903-hclust")
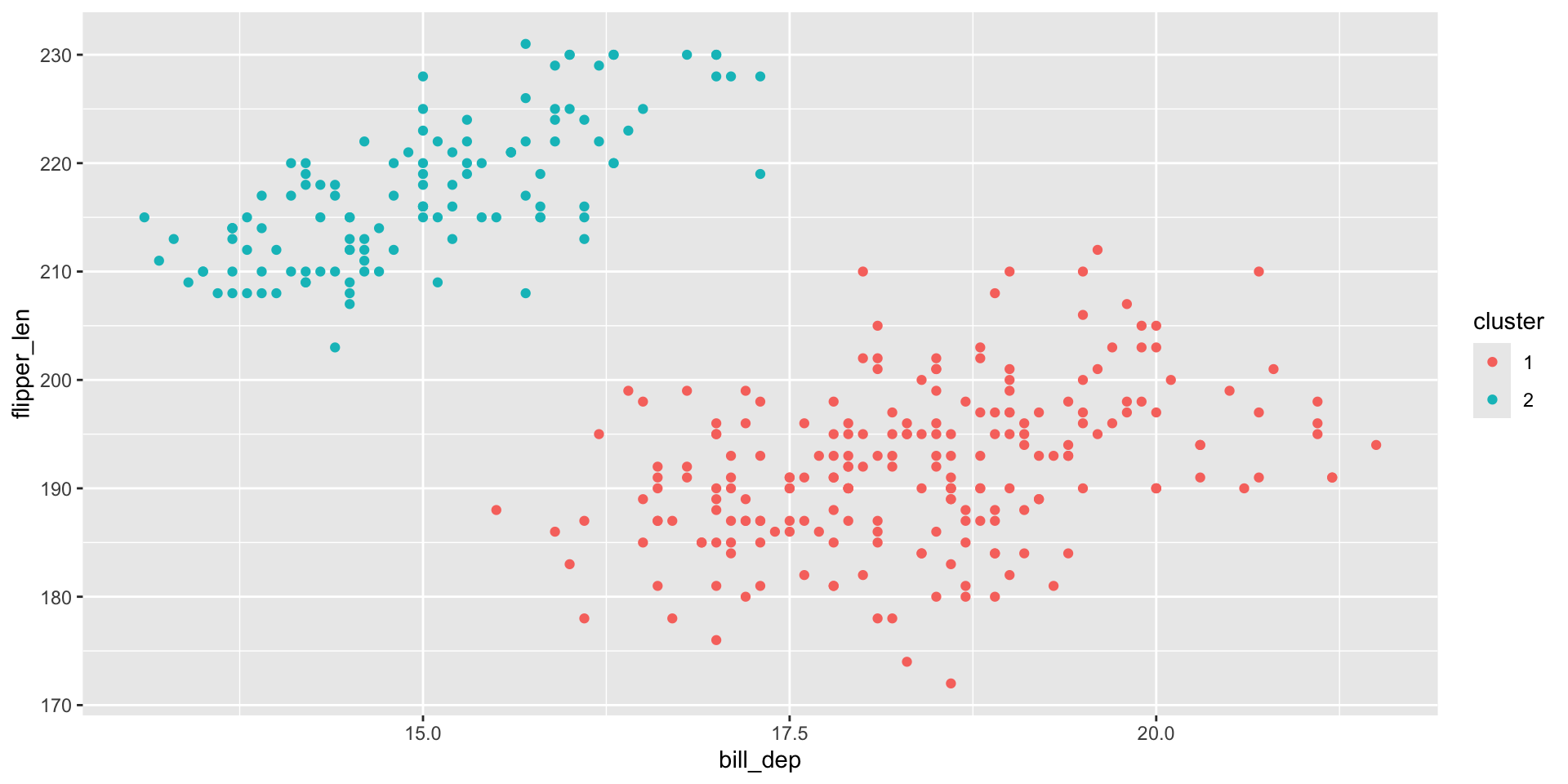
Similar question as in kmeans: do NA values matter?
Hierarchical clustering with the penguins data
To make the illustration simpler, we will take 10 observations for each species:
penguins_small <- penguins_clean |>
group_by(species) |>
slice_head(n = 10) |>
ungroup()
penguins_small# A tibble: 30 × 8
species island bill_len bill_dep flipper_len body_mass sex year
<fct> <fct> <dbl> <dbl> <int> <int> <fct> <int>
1 Adelie Torgersen 39.1 18.7 181 3750 male 2007
2 Adelie Torgersen 39.5 17.4 186 3800 female 2007
3 Adelie Torgersen 40.3 18 195 3250 female 2007
4 Adelie Torgersen 36.7 19.3 193 3450 female 2007
5 Adelie Torgersen 39.3 20.6 190 3650 male 2007
6 Adelie Torgersen 38.9 17.8 181 3625 female 2007
7 Adelie Torgersen 39.2 19.6 195 4675 male 2007
8 Adelie Torgersen 41.1 17.6 182 3200 female 2007
9 Adelie Torgersen 38.6 21.2 191 3800 male 2007
10 Adelie Torgersen 34.6 21.1 198 4400 male 2007
# ℹ 20 more rowsHierarchical clustering
Again, does scaling matter?
# scale the variables
hclust_df <- penguins_small[,3:6] |> scale()
# compute the distance
hclust_dist <- hclust_df |> dist()
# compute the hierarchical clustering
hclust_results1 <- hclust(hclust_dist, method = "single")
hclust_results1
Call:
hclust(d = hclust_dist, method = "single")
Cluster method : single
Distance : euclidean
Number of objects: 30 Visualize hierarchical clustering results
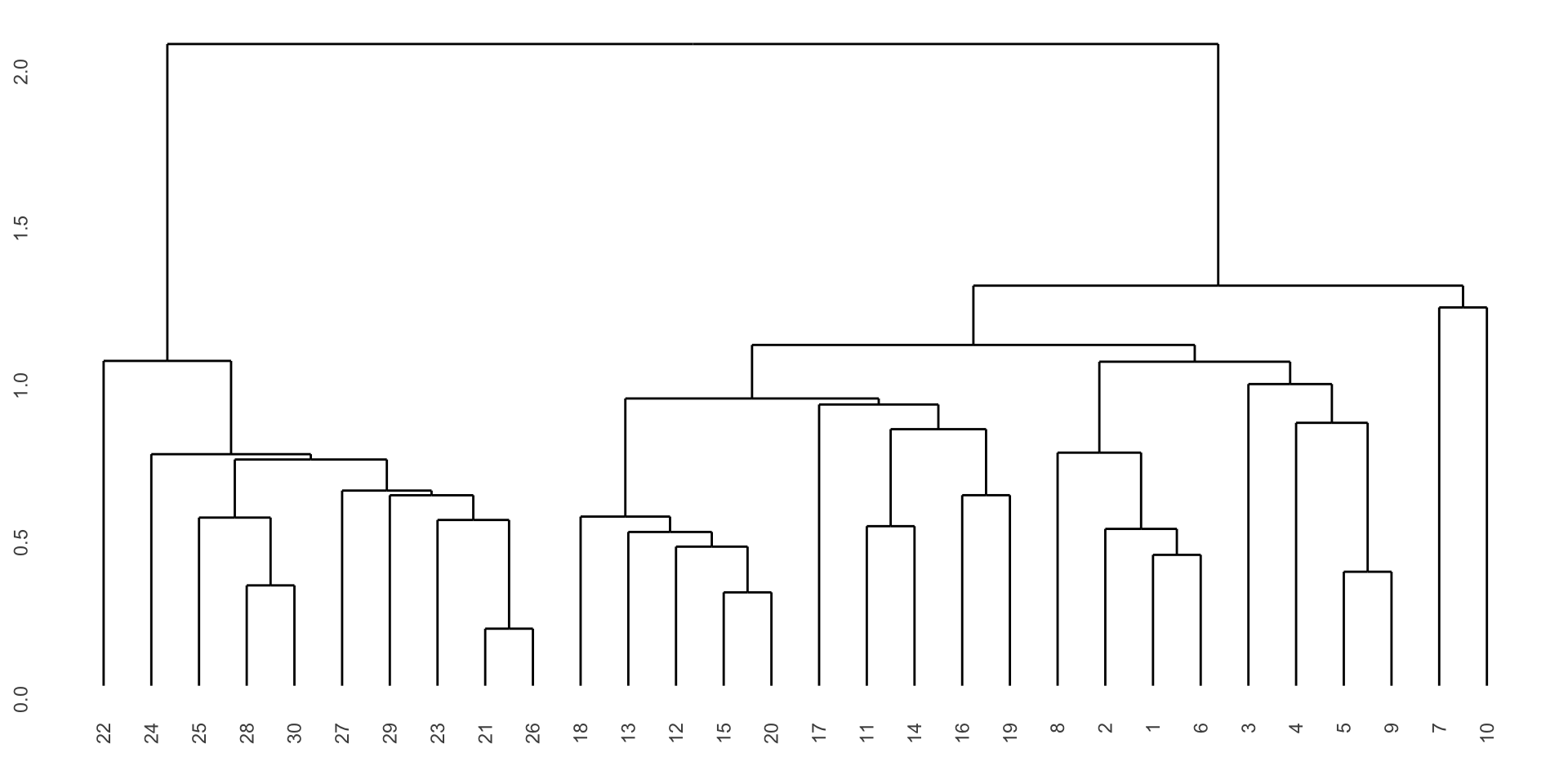
Visualize hierarchical clustering results
Code
hclust_data1 <- ggdendro::dendro_data(hclust_results1)
hclust_segments1 <- as_tibble(hclust_data1$segments)
hclust_labels1 <- as_tibble(hclust_data1$labels) |>
mutate(species = penguins_small$species[as.integer(label)])
p1 <- ggplot() +
geom_segment(data = hclust_segments1,
aes(x = x, y = y, xend = xend, yend = yend)) +
geom_text(data = hclust_labels1,
aes(x = x, y = y - 0.02, label = label,
color = species), vjust = 1, size = 3) +
labs(title = "Single Linkage") +
theme_minimal() +
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
panel.grid = element_blank(),
legend.position = "bottom") +
ylim(-0.1, 2.5)
p1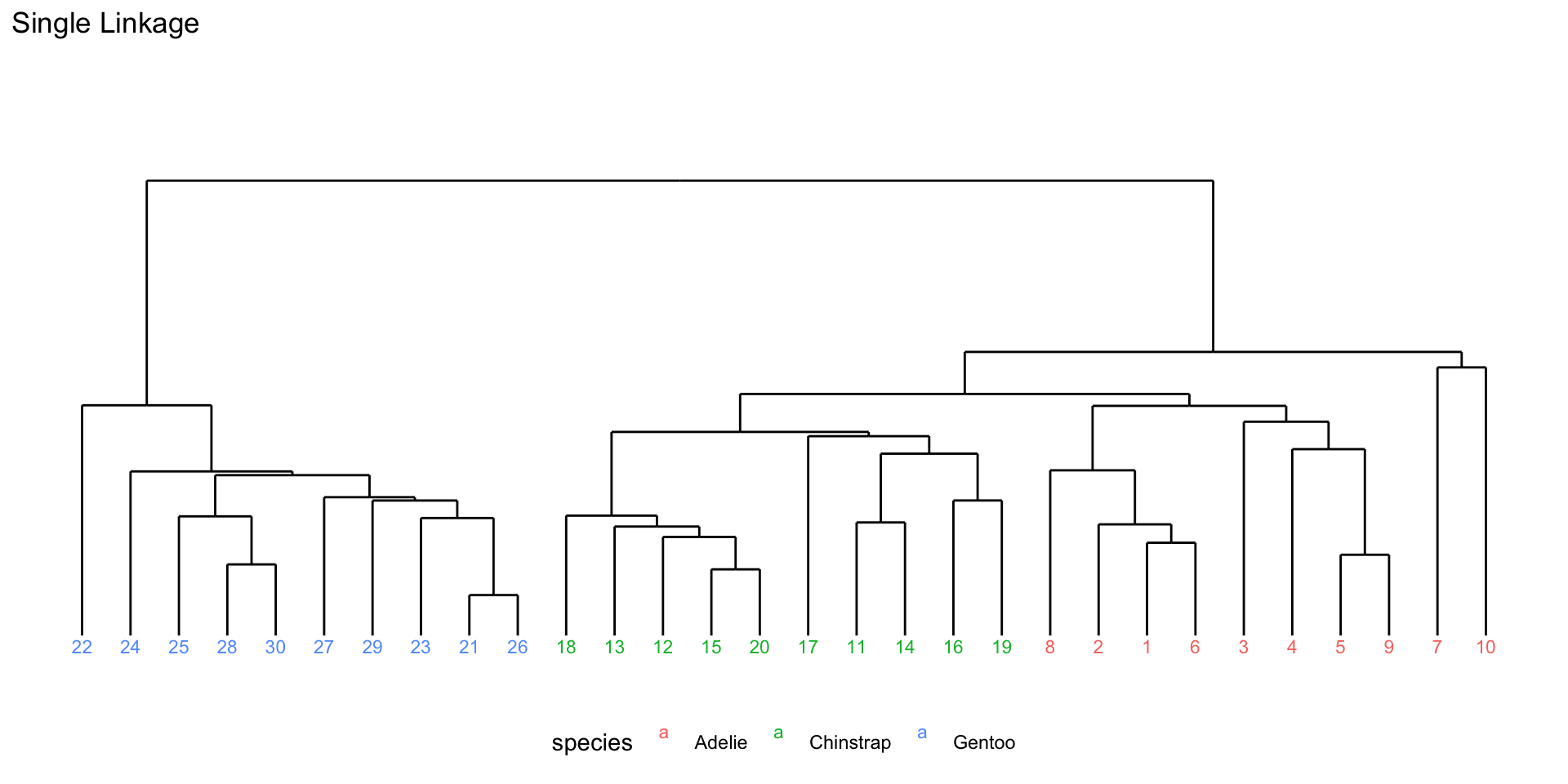
Different linkage methods
- Single linkage: distance between two clusters is defined as the minimum distance between any single member of one cluster and any single member of the other cluster.
- Complete linkage: distance between two clusters is defined as the maximum distance between any single member of one cluster and any single member of the other cluster.
- Average linkage: distance between two clusters is defined as the average distance between all pairs of members from the two clusters.
- Ward’s linkage: distance between two clusters is defined based on the increase in the total within-cluster variance that would result from merging the two clusters.
Selection:
- Single linkage are most popular among statisticians.
- Average and complete linkage are generally preferred over single linkage, as they tend to yield more balanced dendrograms
Different linkage for penguins data
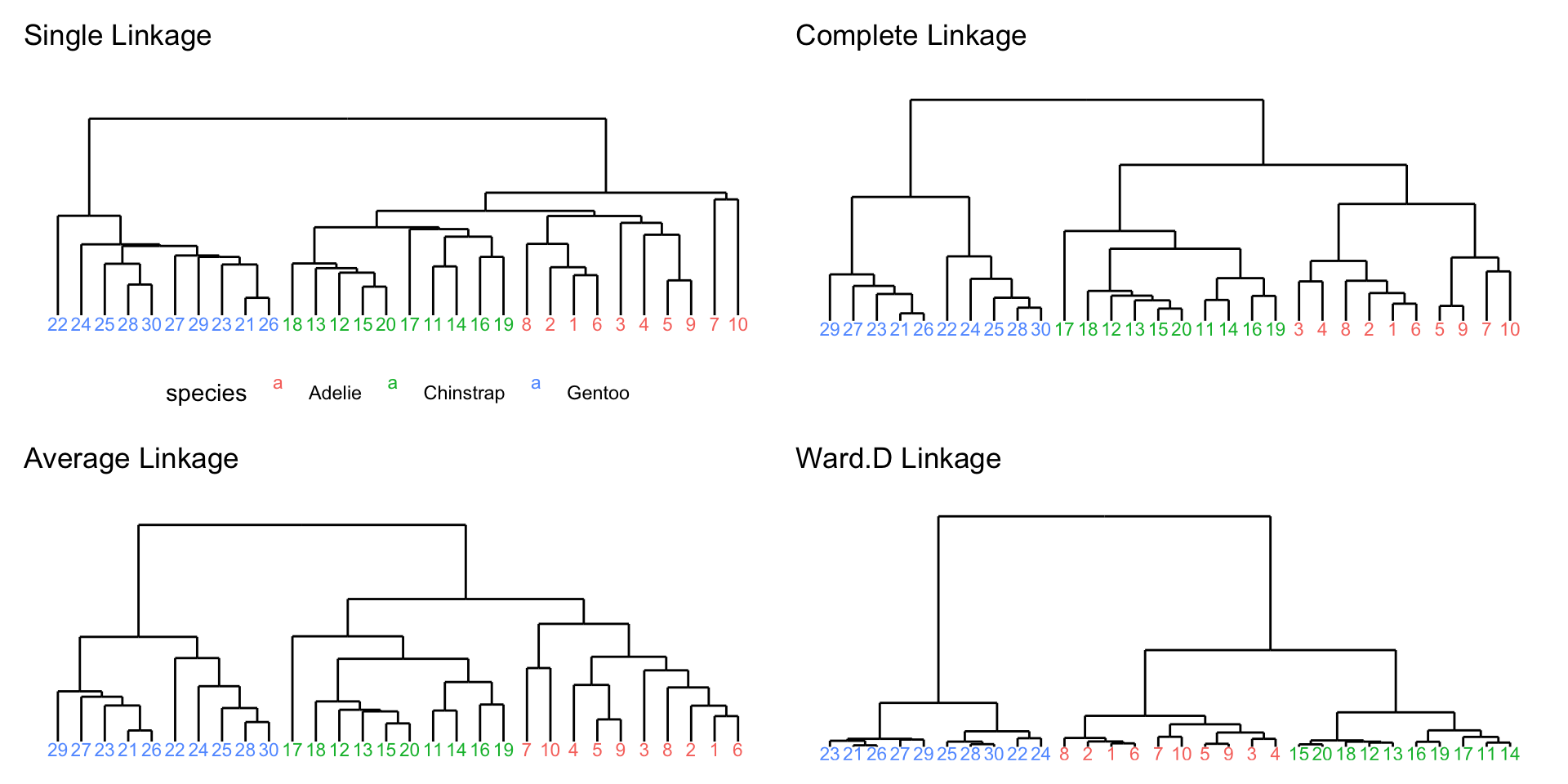
Full penguins data
hclust_df <- penguins_clean[,3:6] |> scale()
# compute the distance
hclust_dist <- hclust_df |> dist()
# compute the hierarchical clustering
hclust_results <- hclust(hclust_dist, method = "average")
hclust_results
Call:
hclust(d = hclust_dist, method = "average")
Cluster method : average
Distance : euclidean
Number of objects: 333 Full penguins data
Code
hclust_data <- ggdendro::dendro_data(hclust_results)
hclust_segments <- as_tibble(hclust_data$segments)
hclust_labels <- as_tibble(hclust_data$labels) |>
mutate(species = penguins_clean$species[as.integer(label)])
ggplot() +
geom_segment(data = hclust_segments,
aes(x = x, y = y, xend = xend, yend = yend)) +
geom_text(data = hclust_labels,
aes(x = x, y = y - 0.02, label = label,
color = species), vjust = 1, size = 3) +
labs(title = "Average Linkage") +
theme_minimal() +
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
panel.grid = element_blank(),
legend.position = "none") +
ylim(-0.1, 4) 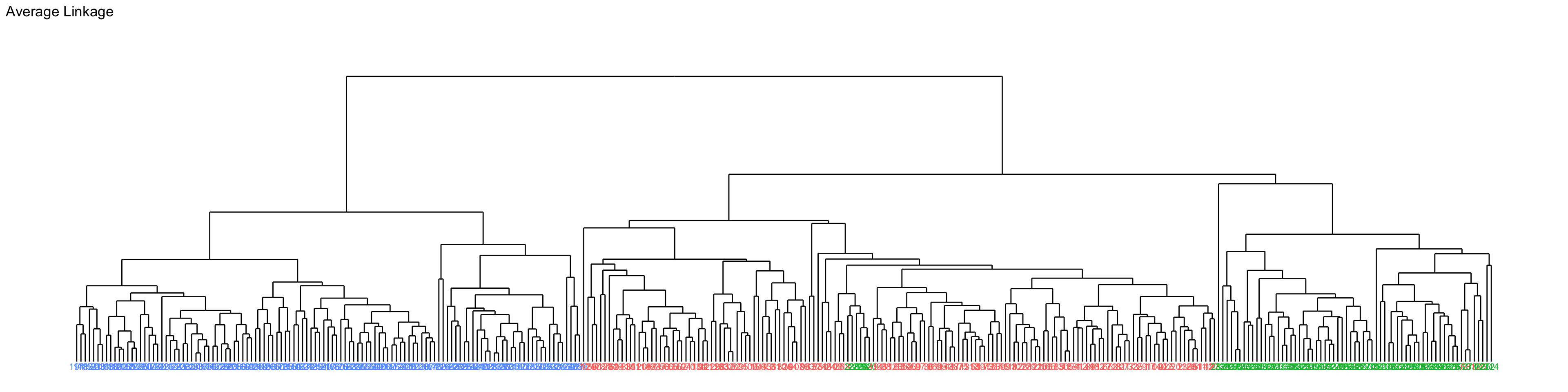
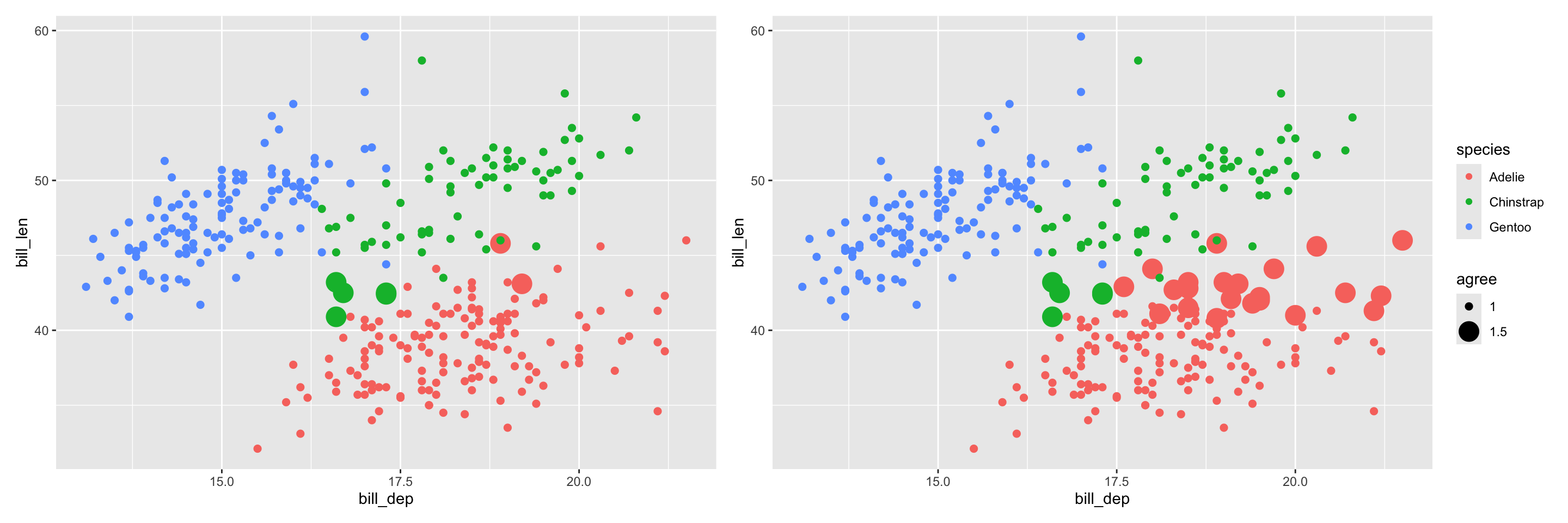
Cut the tree into two clusters
The base R function cutree() can be used to cut the dendrogram into a specified number of clusters.
Compare hierarchical clustering with kmeans
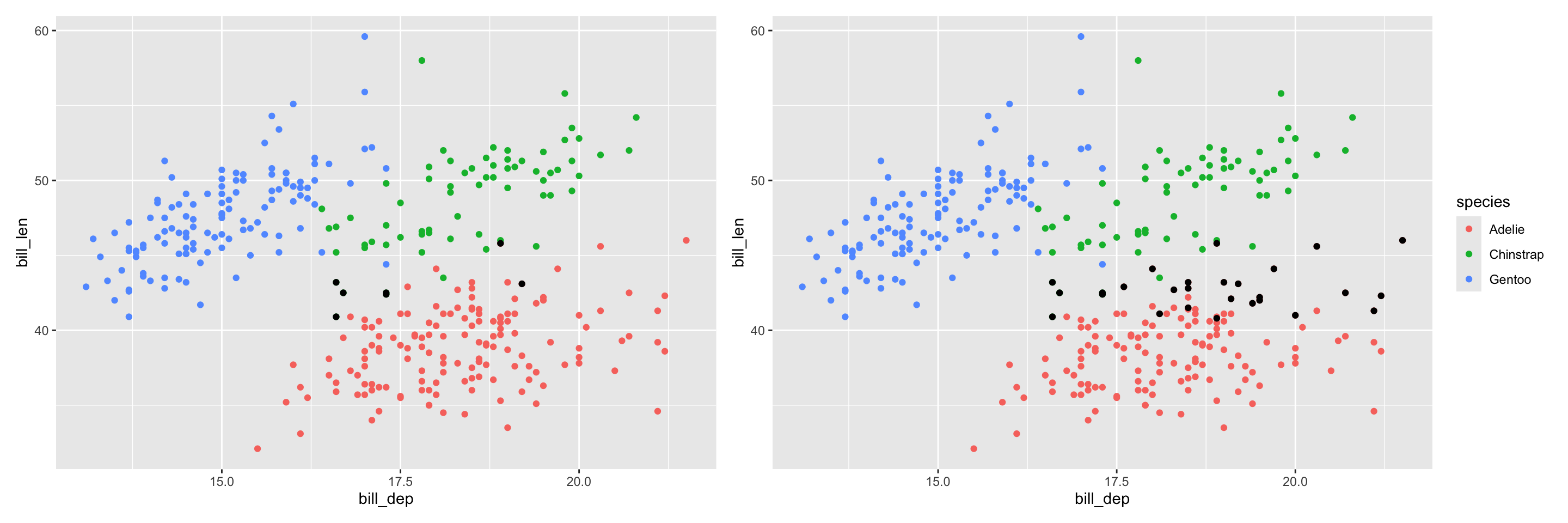
Hierarchical clustering: cut into 3 groups
# A tibble: 5 × 3
cluster species n
<fct> <fct> <int>
1 1 Adelie 144
2 1 Chinstrap 5
3 2 Adelie 2
4 2 Chinstrap 63
5 3 Gentoo 119Kmeans clustering: choose 3 centers
# A tibble: 5 × 3
cluster species n
<fct> <fct> <int>
1 1 Adelie 124
2 1 Chinstrap 5
3 2 Adelie 22
4 2 Chinstrap 63
5 3 Gentoo 119