Elements of Data Science
SDS 322E
Department of Statistics and Data Sciences
The University of Texas at Austin
Fall 2025
Learning objectives
- Understand the basic idea of decision trees for regression and classification
- Fit decision trees using
tidymodelsfor both regression and classification tasks - Identify the key parameters of decision trees and their effects
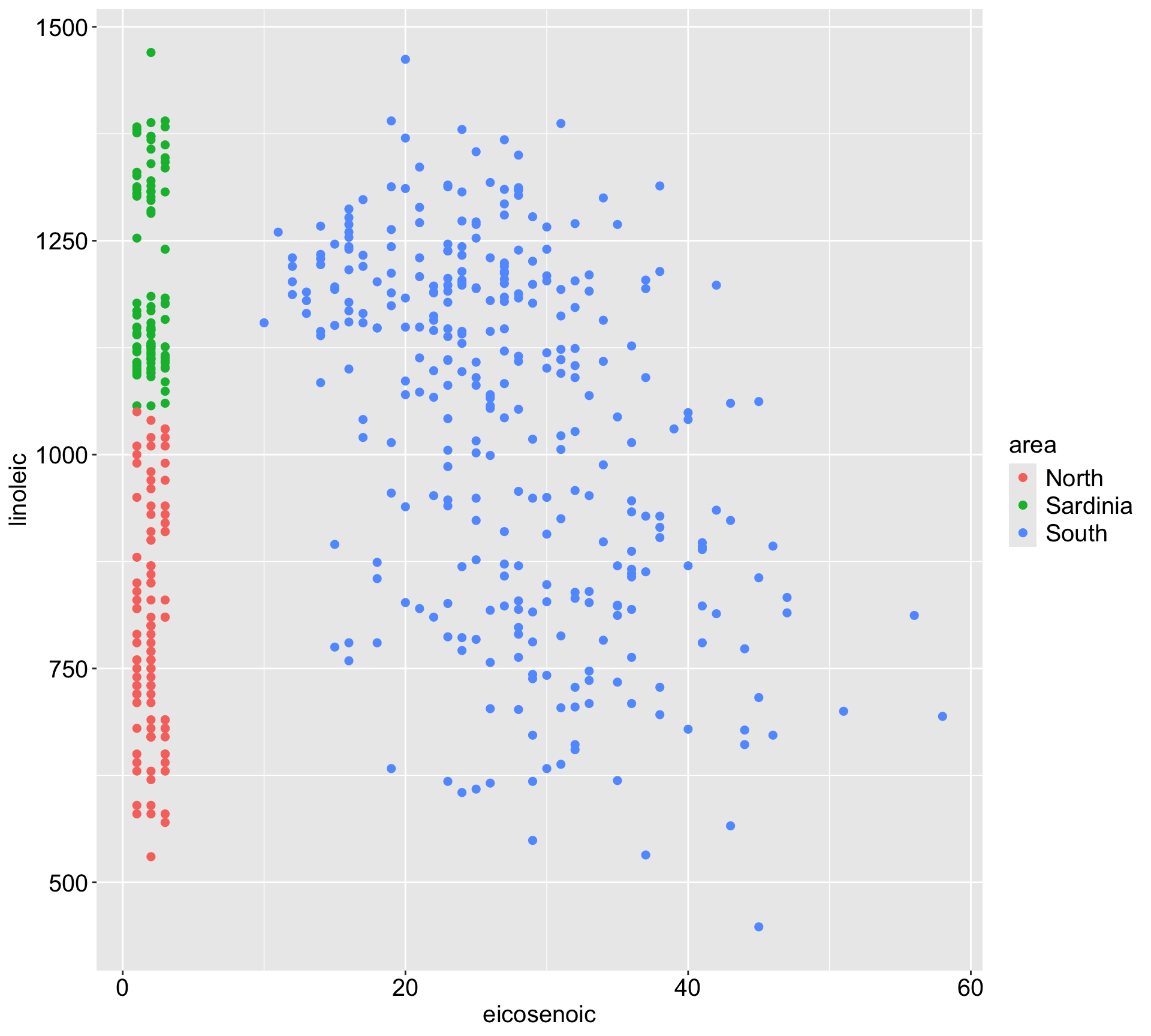
If we would like to predict the area from x1 (eicosenoic) and x2 (linoleic), would you expect linear regression or KNN to perform well?
Decision rules
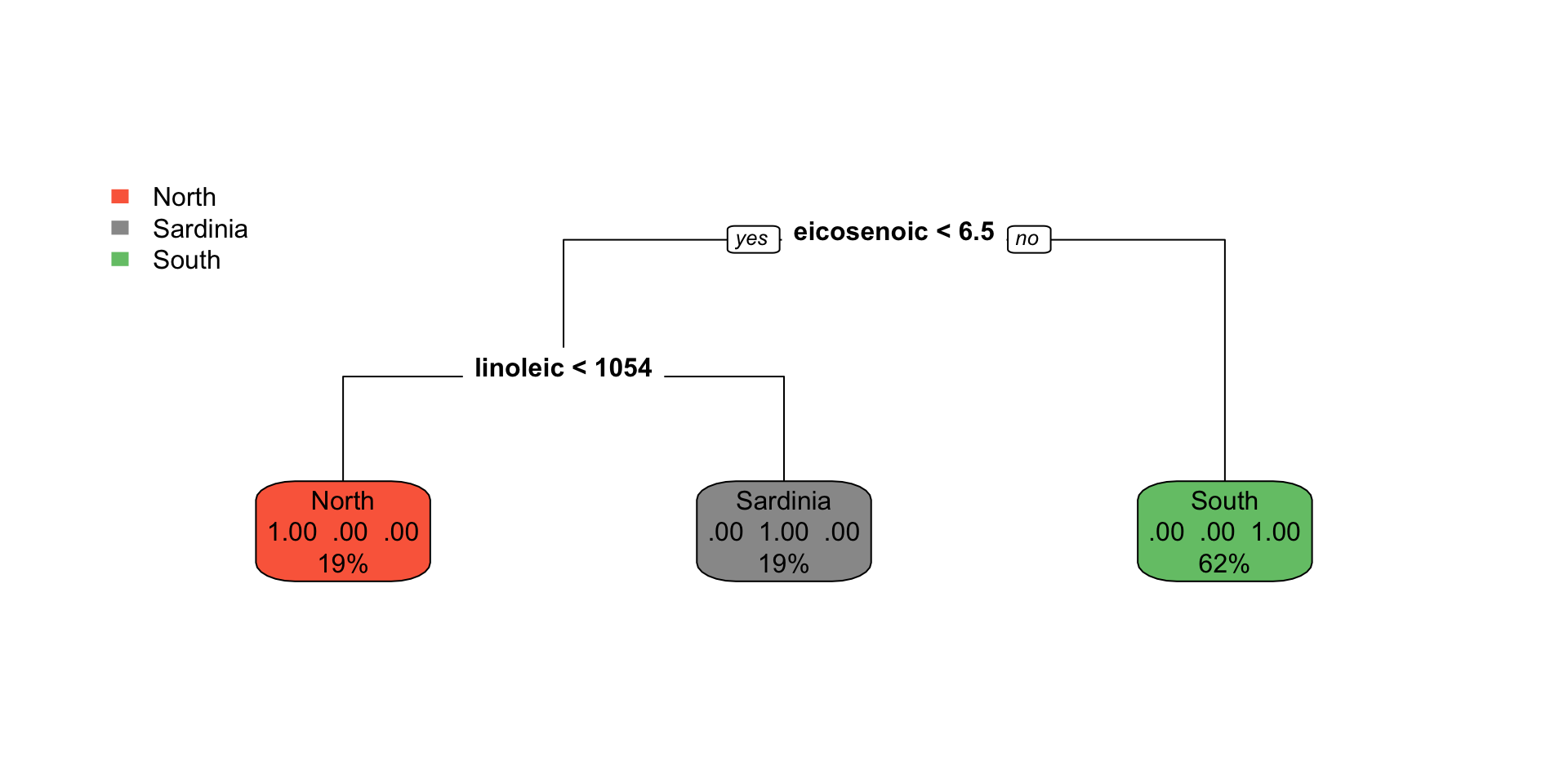
Regression tree
Divide/partition the predictor space \(X_1, \ldots, X_p\) into \(J\) regions \(R_1, \ldots, R_J\).
For every observation that falls within region \(R_j\), we make the same prediction \(\bar{y}_{R_j}\), which is the mean of the response values for the training observations in \(R_j\).
For regression, the overall goal is typically to minimize the residual sum of squares:\[ RSS = \sum_{j=1}^{J} \sum_{i \in R_j} (y_i - \bar{y}_{R_j})^2\]
Regression tree with tidymodels
Step 1/2/3: specify the model/ recipe/ workflow:
Step 4: fit the tree:
Step 5/6: predict/ calculate the prediction accuracy metrics:
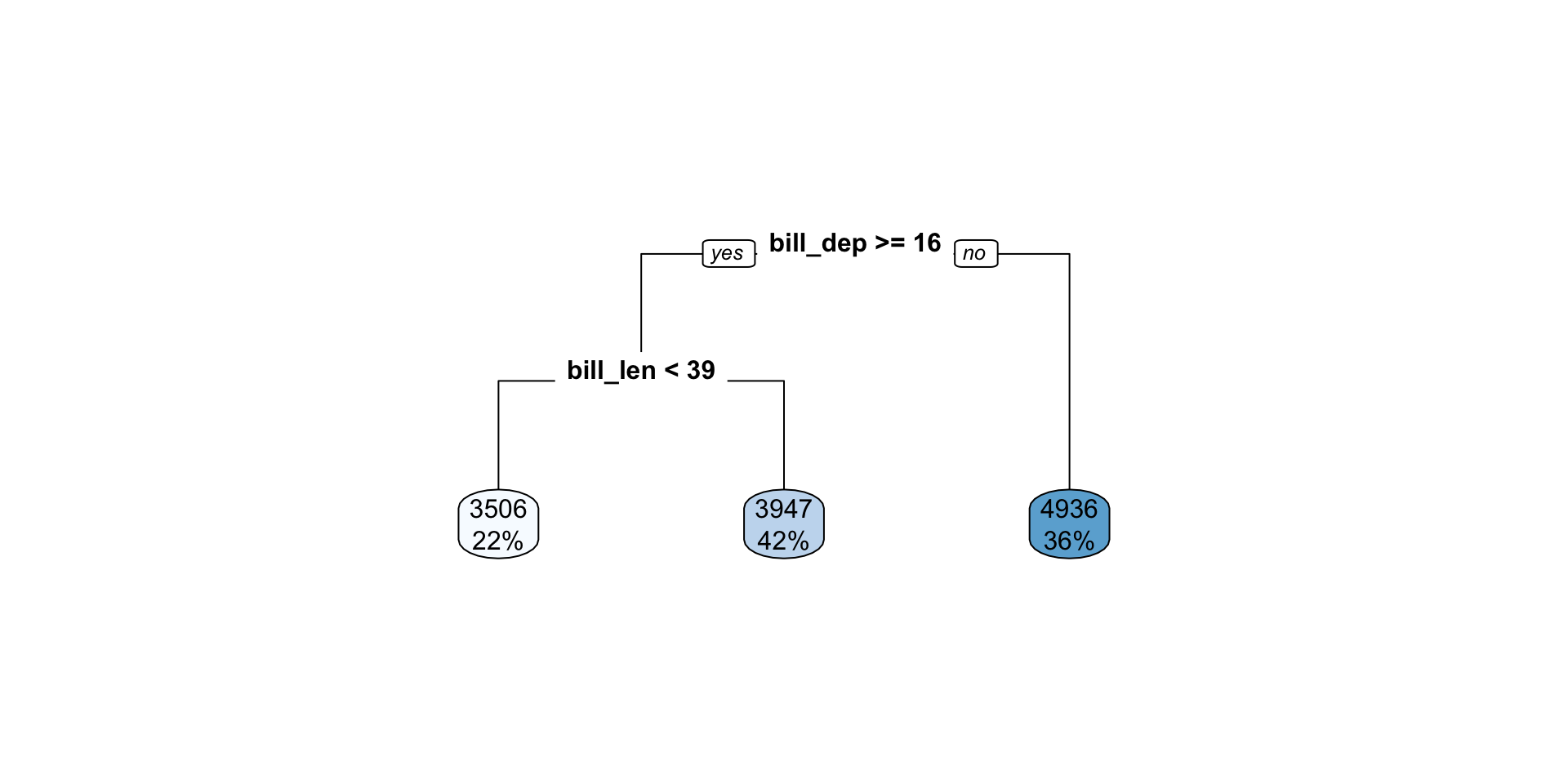
Regression tree with tidymodels
n=342 (2 observations deleted due to missingness)
node), split, n, deviance, yval
* denotes terminal node
1) root 342 219307700 4201.754
2) bill_dep>=16.45 220 65255400 3794.318
4) bill_len< 39.05 76 9524836 3505.921 *
5) bill_len>=39.05 144 46073260 3946.528 *
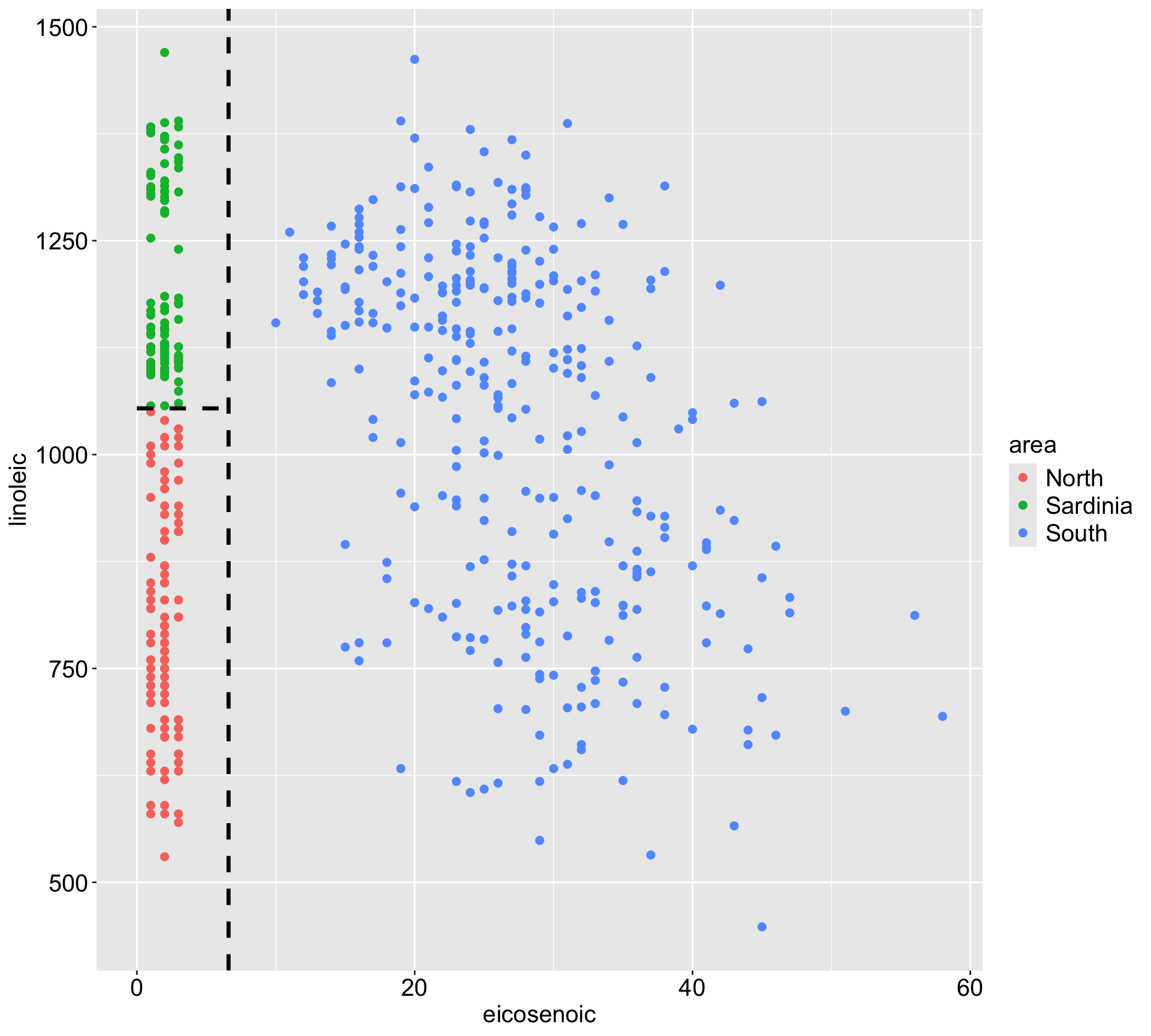
3) bill_dep< 16.45 122 51673930 4936.475 *Visualize the space
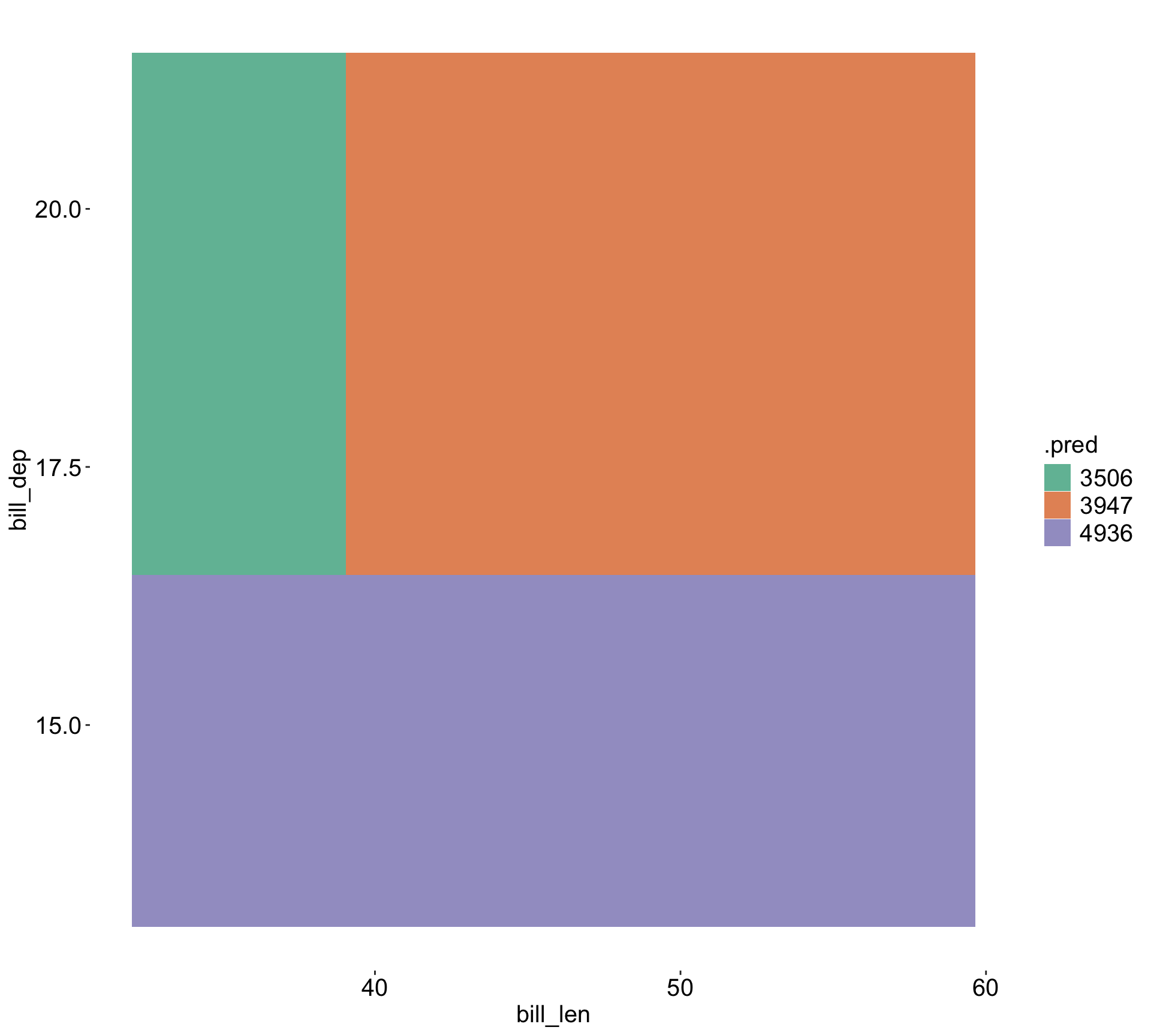
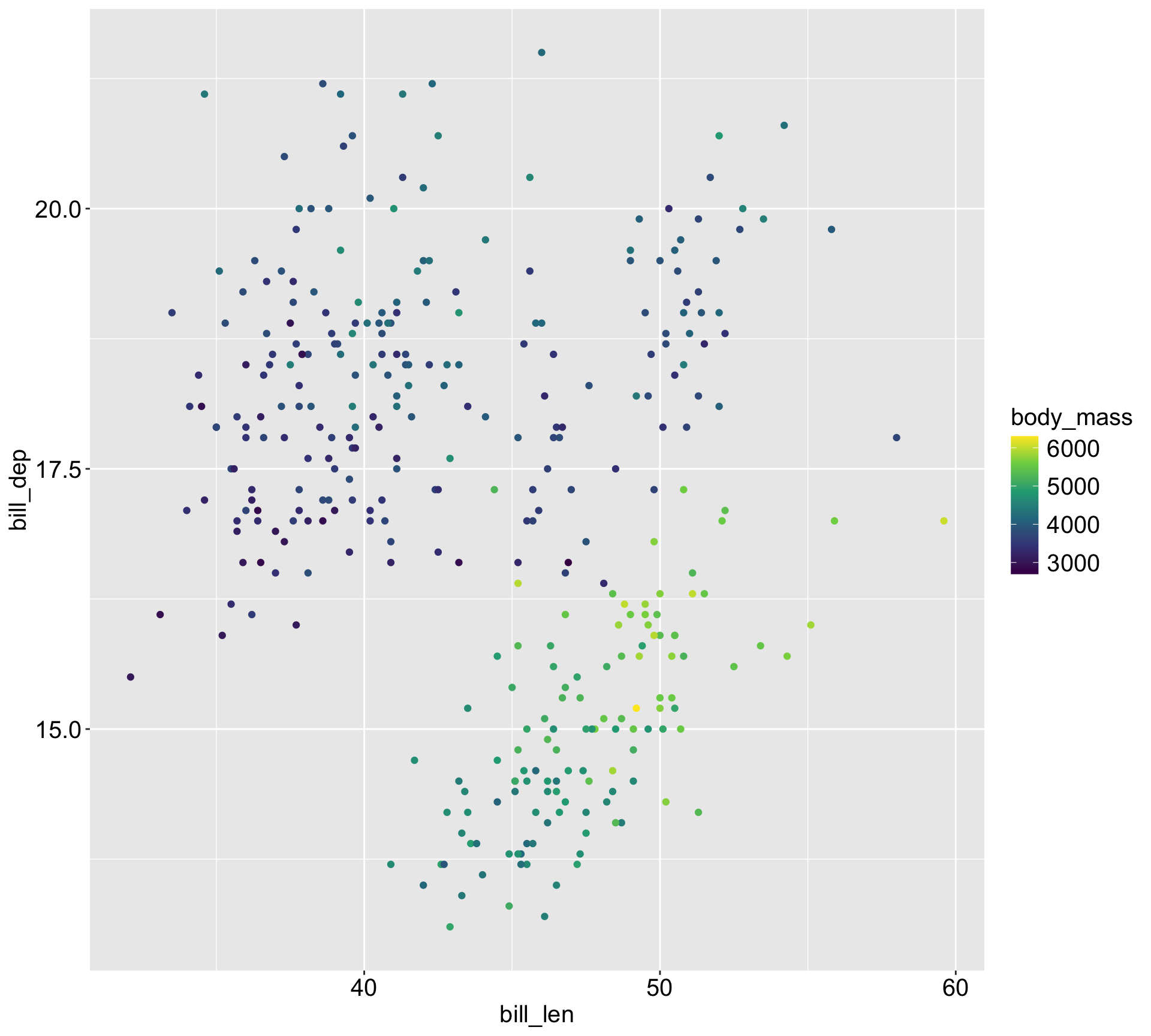
| Species | Adelie | Chinstrap | Gentoo |
|---|---|---|---|
| Average body mass | 3701 | 3733 | 5076 |
Classification tree
- Classification trees use the same splits, but minimize impurity (not RSS).
- Classification of an observation is taken as the majority class in a region
- Goal is to increase the “purity” of each region so that most regions are dominated by a single class

Classification tree
Let \(p_{mk}\) be the proportion of training observations in region \(R_m\) that are from class \(k\). For region \(R_m\),
The Gini index: \(G(R_m) = \sum_{k=1}^{K} p_{mk}(1 - p_{mk})\). lower, purer.
The Entropy: \(H(R_m) = - \sum_{k=1}^{K} p_{mk} \log(p_{mk})\). lower, purer.
Example:
After a certain split, we have 10 observations in region \(R_m\), with
| class 1 | class 2 | class3 |
|---|---|---|
| 4 | 3 | 3 |
Then, we may calculate:
- Gini index: \(G(R_m) = 0.4 *(1- 0.4) + 0.3 * (1-0.3) + 0.3 * (1-0.3) = 0.66\)
- Entropy: \(H(R_m) = - (0.4 \log 0.4 + 0.3 \log 0.3 + 0.3 \log 0.3) = 1.09\)
Here we predict class 1 since it is the majority vote.
Classification tree
The next cut may be dividing these 10 observations into two regions:
- \(R_{m1}\) with 6 observations (4 from class 1, 2 from class 2) and
- \(R_{m2}\) with 4 observations (1 from class 2, 3 from class 3).
Then we would predict class 1 for \(R_{m1}\) and class 3 for \(R_{m2}\).
Gini index:
- \(G(R_{m1}) = 4/6 * (1-4/6) + 2/6 * (1- 2/6) = 0.44\),
- \(G(R_{m2}) = 1/4 * (1 - 1/4) + 3/4 * (1-3/4) = 0.375\)
Entropy:
- \(H(R_{m1}) = -(4/6 * log(4/6) + (2/6) *log(2/6)) = 0.64\),
- \(H(R_{m2}) = - (1/4 * log(1/4) + 3/4 * log(3/4) = 0.56\)
Why does purity matter?
Example: Classification tree with tidymodels
Step 1/2/3: Setup the recipe/ model/ workflow:
Step 4: Fit
Step 5: Predict/ Calculate classification metric
Classification tree with tidymodels
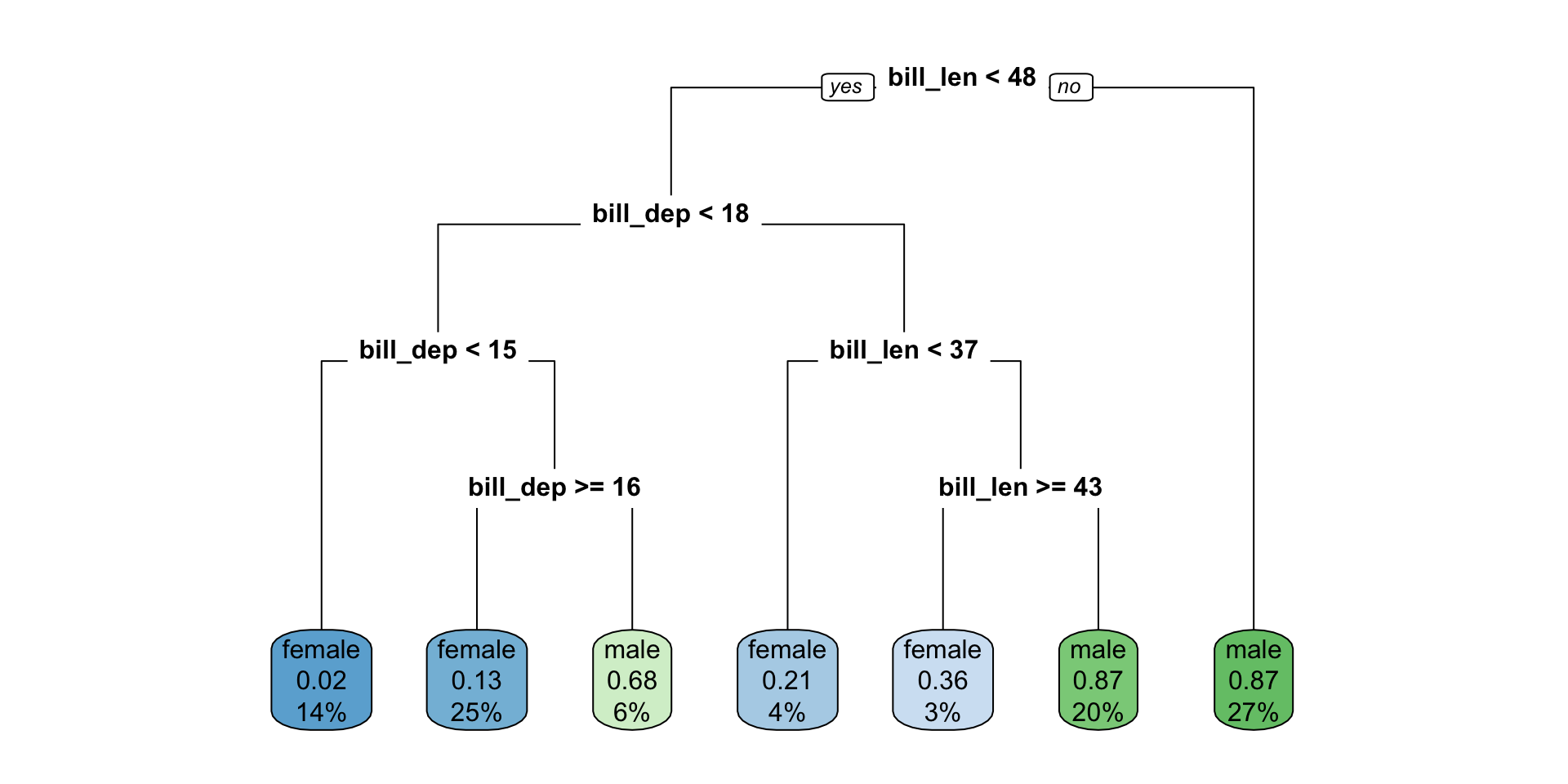
Visualize the space
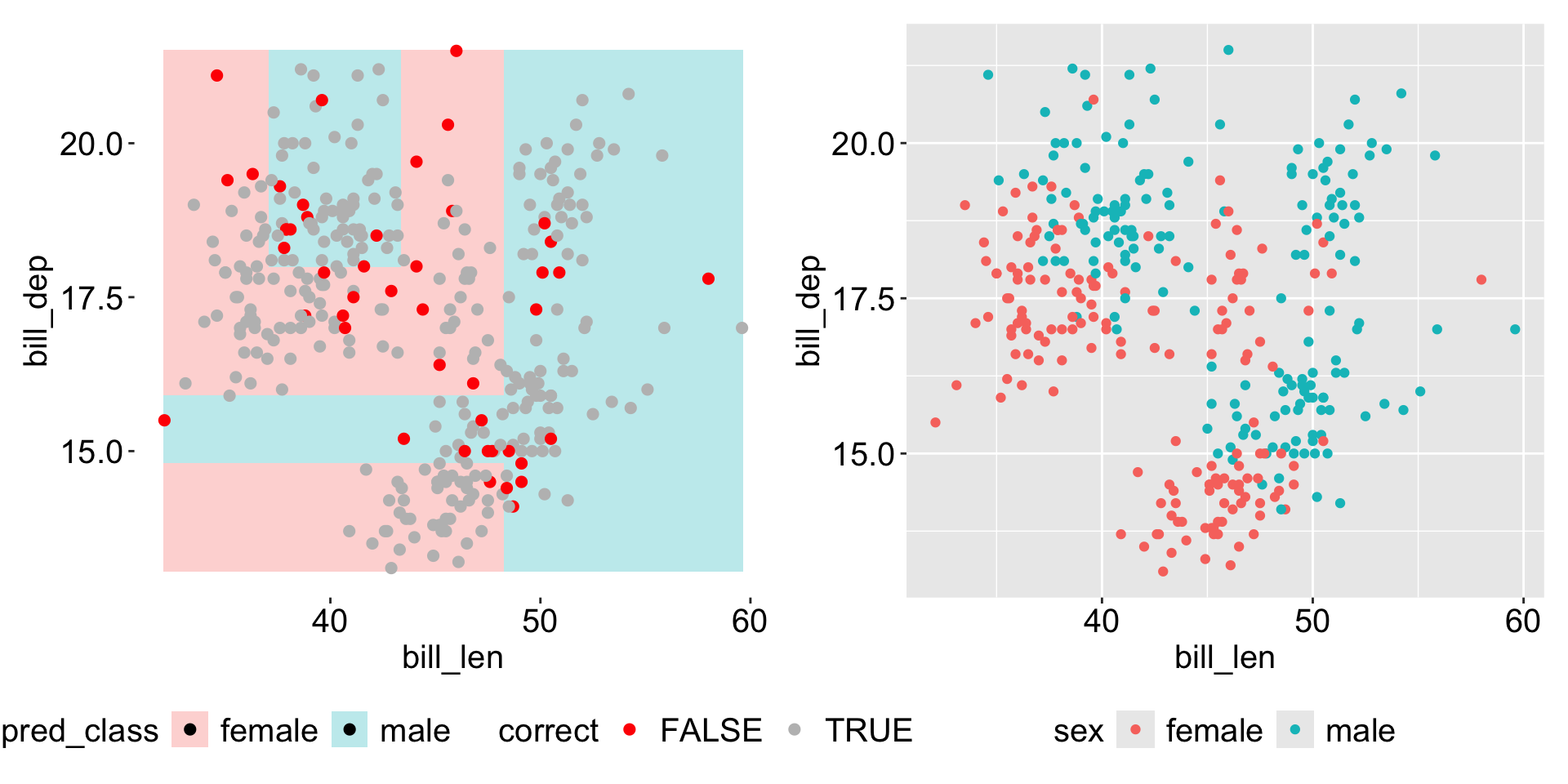
More details on the tree parameters
- By default, the Gini index is used for splitting the tree, to change to entropy, use
Trees have a few hyper-parameters that you may want to tune:
min_n: minimum number of data points in a node required for the node to be split furthercost_complexity: complexity parameter that penalizes large treestree_depth: maximum depth of the tree