Elements of Data Science
SDS 322E
Department of Statistics and Data Sciences
The University of Texas at Austin
Fall 2025
Assessment heads-up
- When completing your assessment, remember to use line breaks on your code. For example,
is better than
Make sure when you knit a pdf, the code is not cut off. You will lose marks if we can’t see your code.
- There are only 8 questions in your homework 1. Gradescope may ask you to crop for Question 9, but we don’t have a Question 9. Just ignore it.
Learning objectives
We will
compare a
data.framevs. atibble(preferred)use the
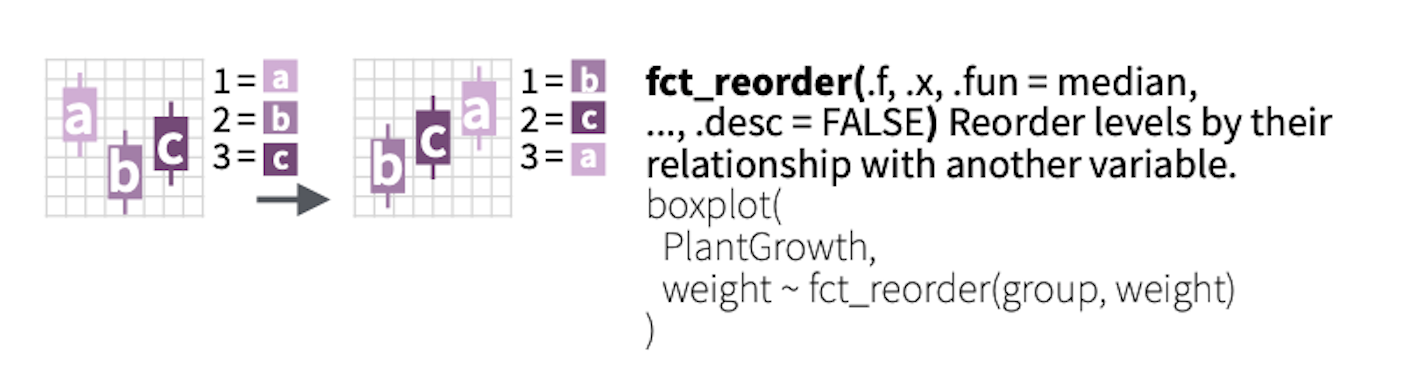
forcatspackage to manipulate factor variablesas_factor(),fct_reorder(), andfct_recode()
change legend title in the plot with
labs()dive deeper into color use in data visualization
- What’s a good vs. bad color palette? Examples: viridis vs. rainbow
- perception uniformity
- Colorblindness friendliness
- Qualitative, Sequential, and diverging palettes
- A new package to explore:
ggthemes
- What’s a good vs. bad color palette? Examples: viridis vs. rainbow
Data frame vs. tibble
Data frame:
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2Tibble:
# A tibble: 32 × 11
mpg cyl disp hp drat wt qsec vs am gear carb
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
# ℹ 27 more rowsA few things that are inconvenient with a data frame:
A data frame will print all the rows, while a tibble only prints the first 10 rows. You will need to scroll all the way up to see the column names in a data frame.
A tibble has a few prints that make it easier to know your data, e.g. 1) data dimension: 32 x 11, 2) variable type:
<dbl>
Convert a data frame to a tibble
For some historical reasons, a data frame allows you to specify a rowname:
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1There is a convenient function to make it as a variable in a tibble:
Then convert it to a tibble:
# A tibble: 3 × 12
model mpg cyl disp hp drat wt qsec vs am gear carb
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Mazda RX4 21 6 160 110 3.9 2.62 16.5 0 1 4 4
2 Mazda RX4 W… 21 6 160 110 3.9 2.88 17.0 0 1 4 4
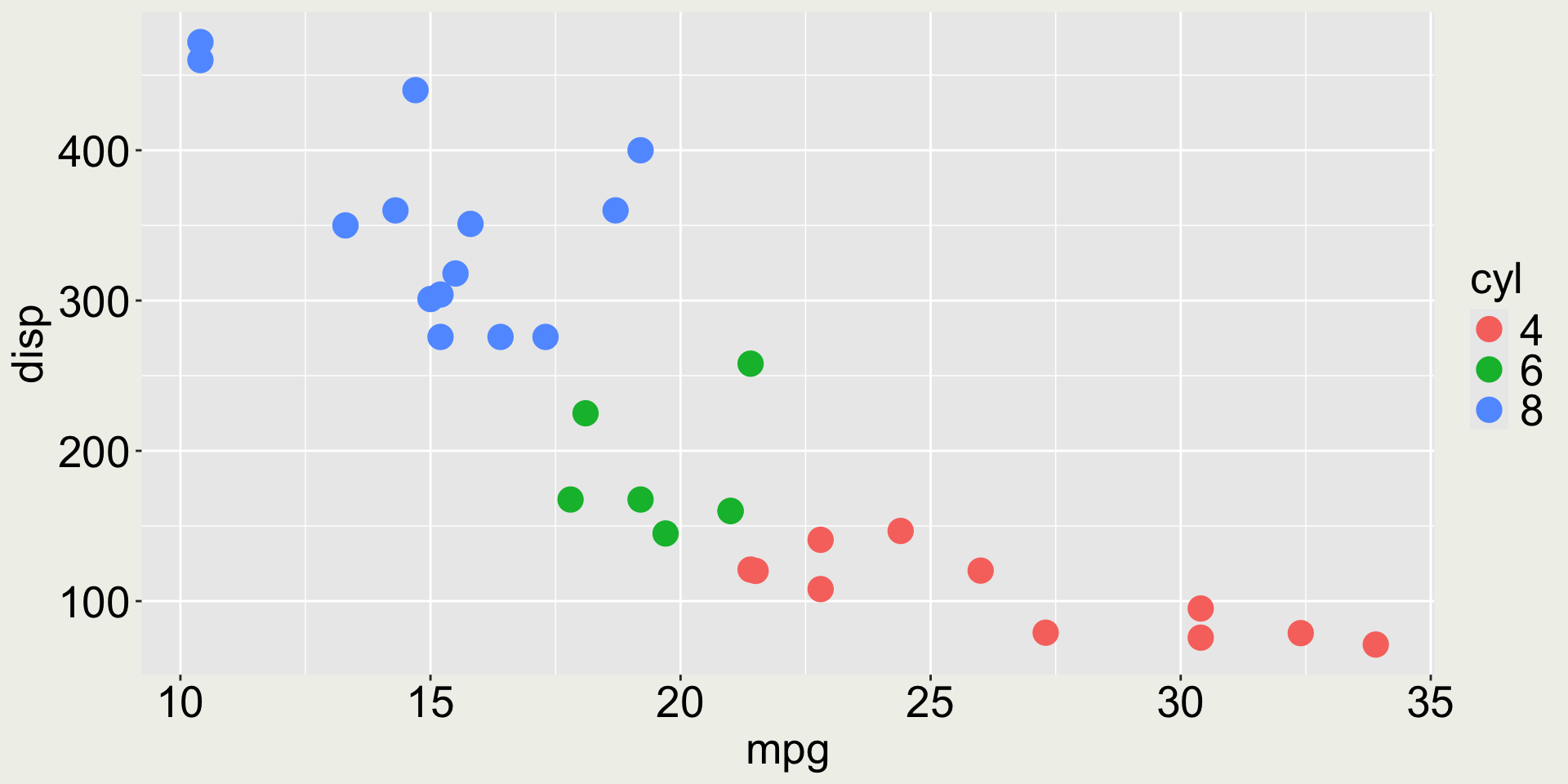
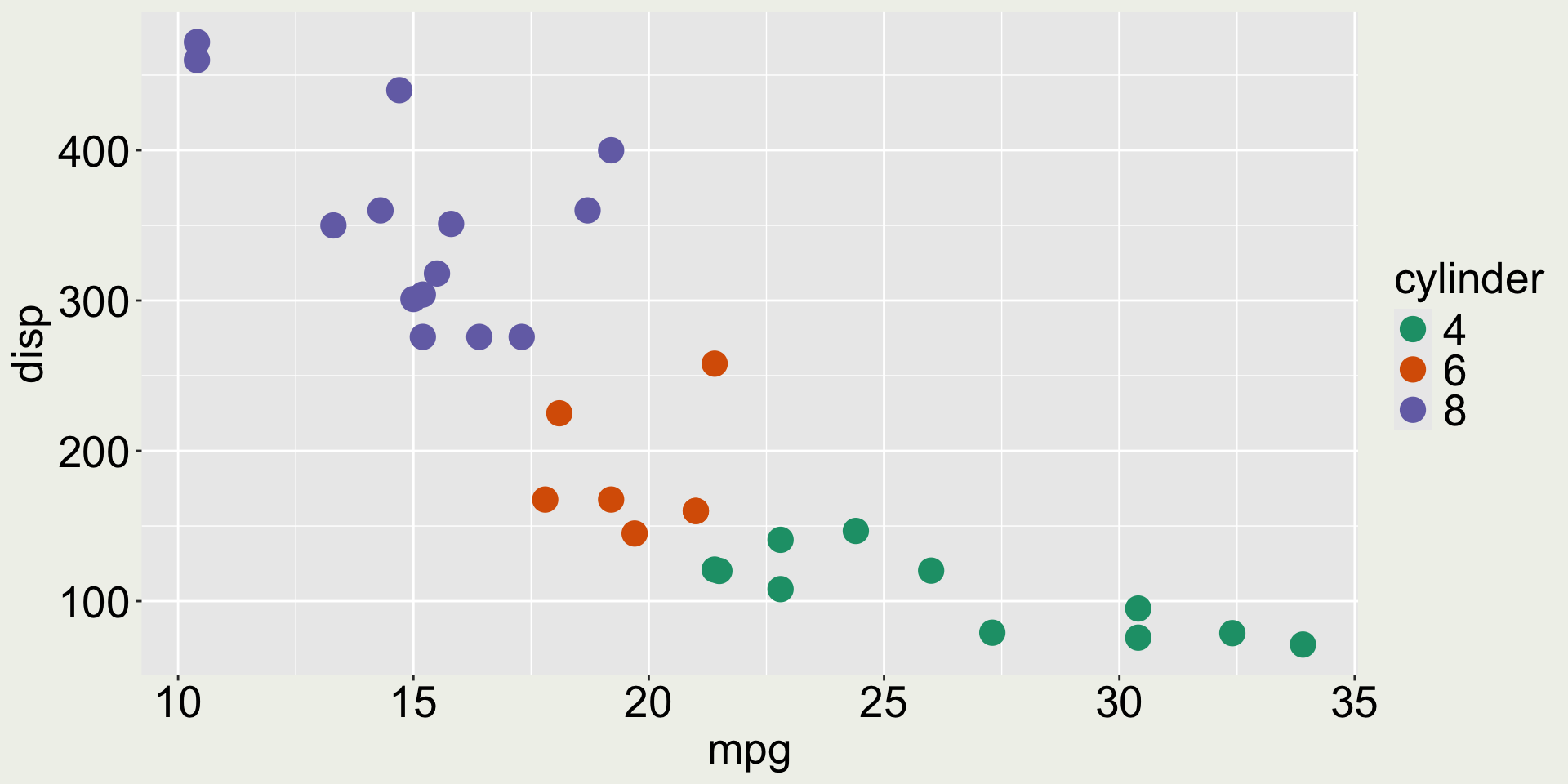
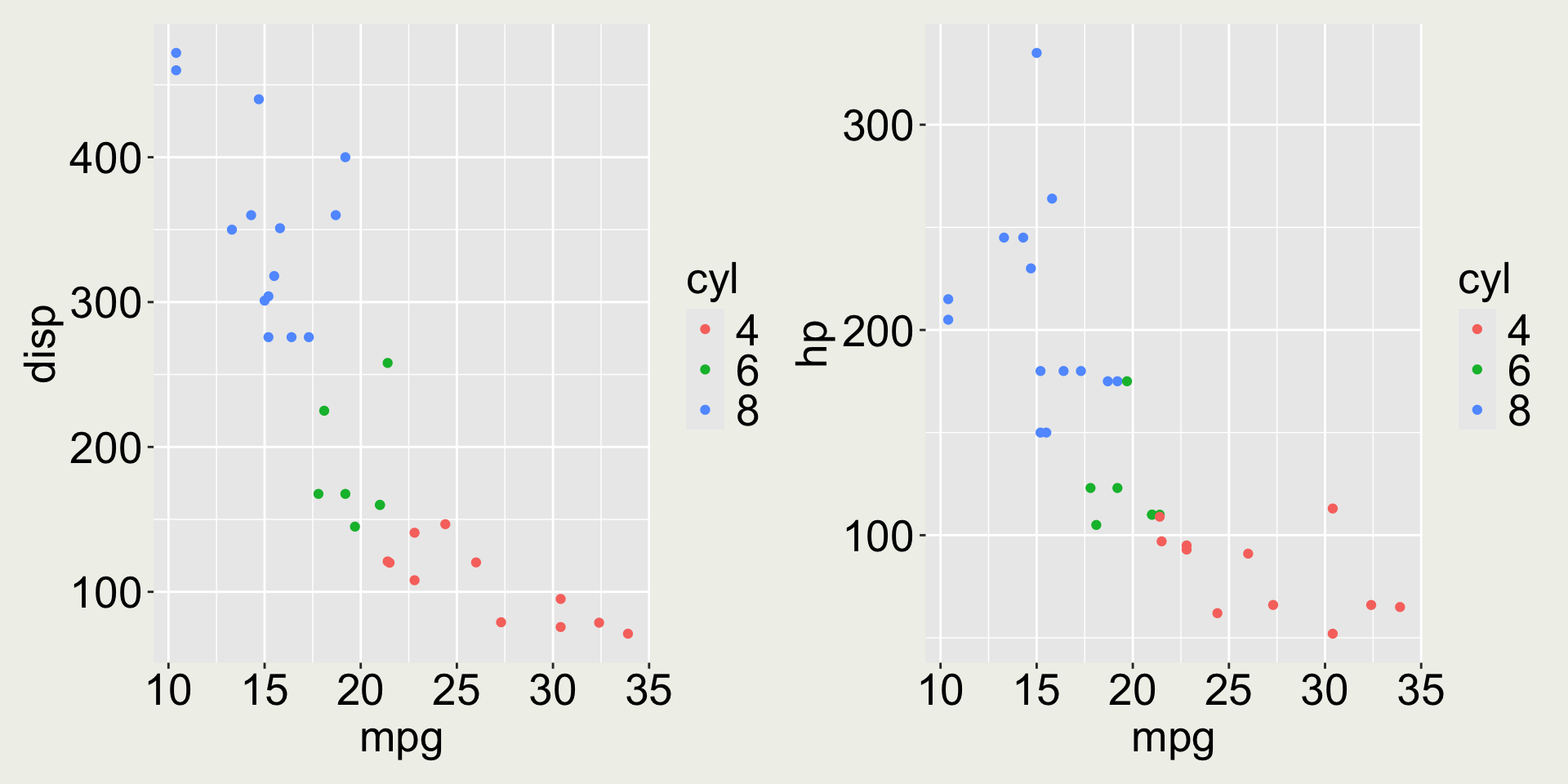
3 Datsun 710 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1Anything wrong with the color in this plot?
mtcars: mpg vs. disp colored by cyl
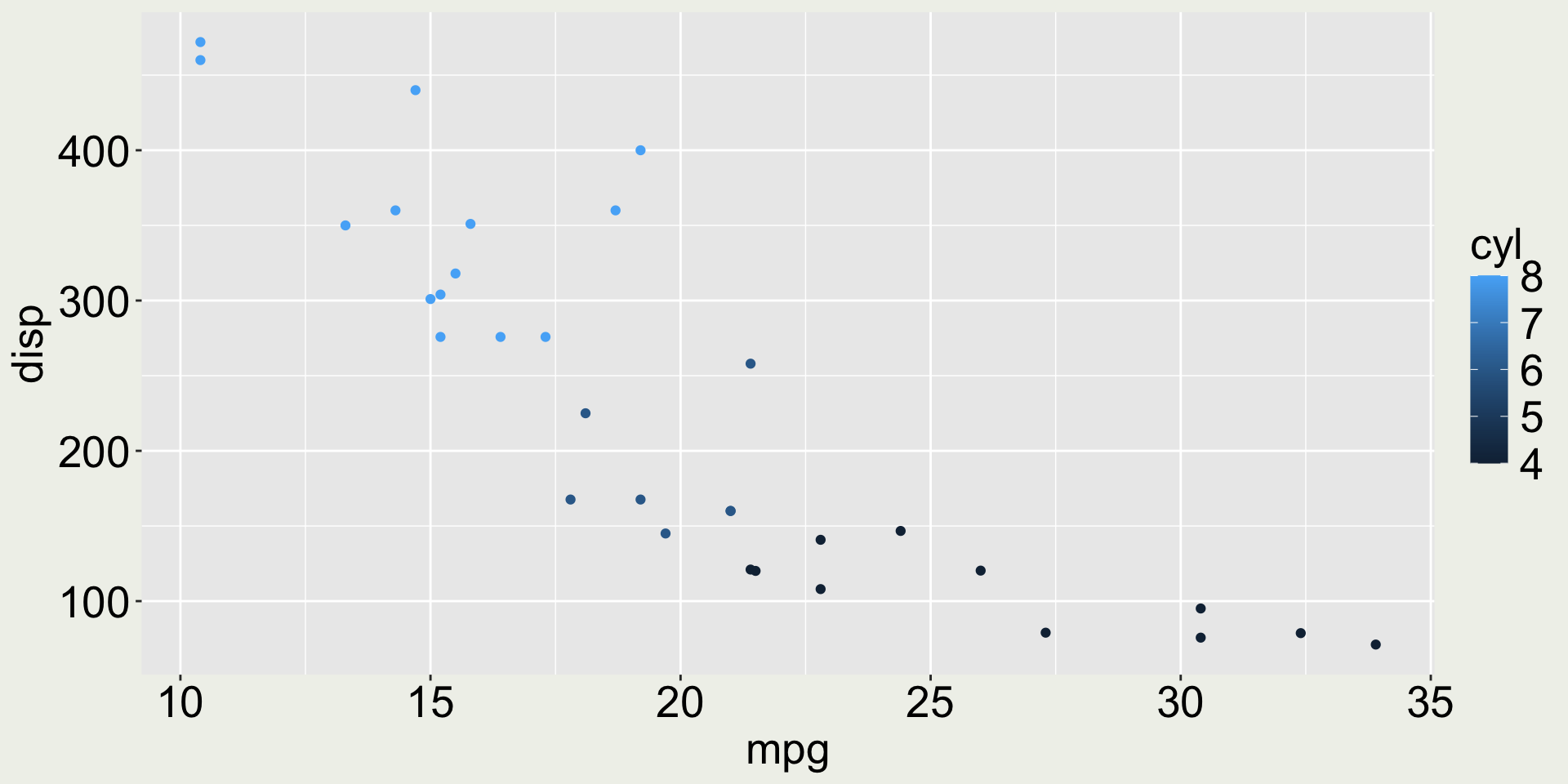
The variable cyl only has three values (4, 6, 8) but it is mapped to a continuous scale.
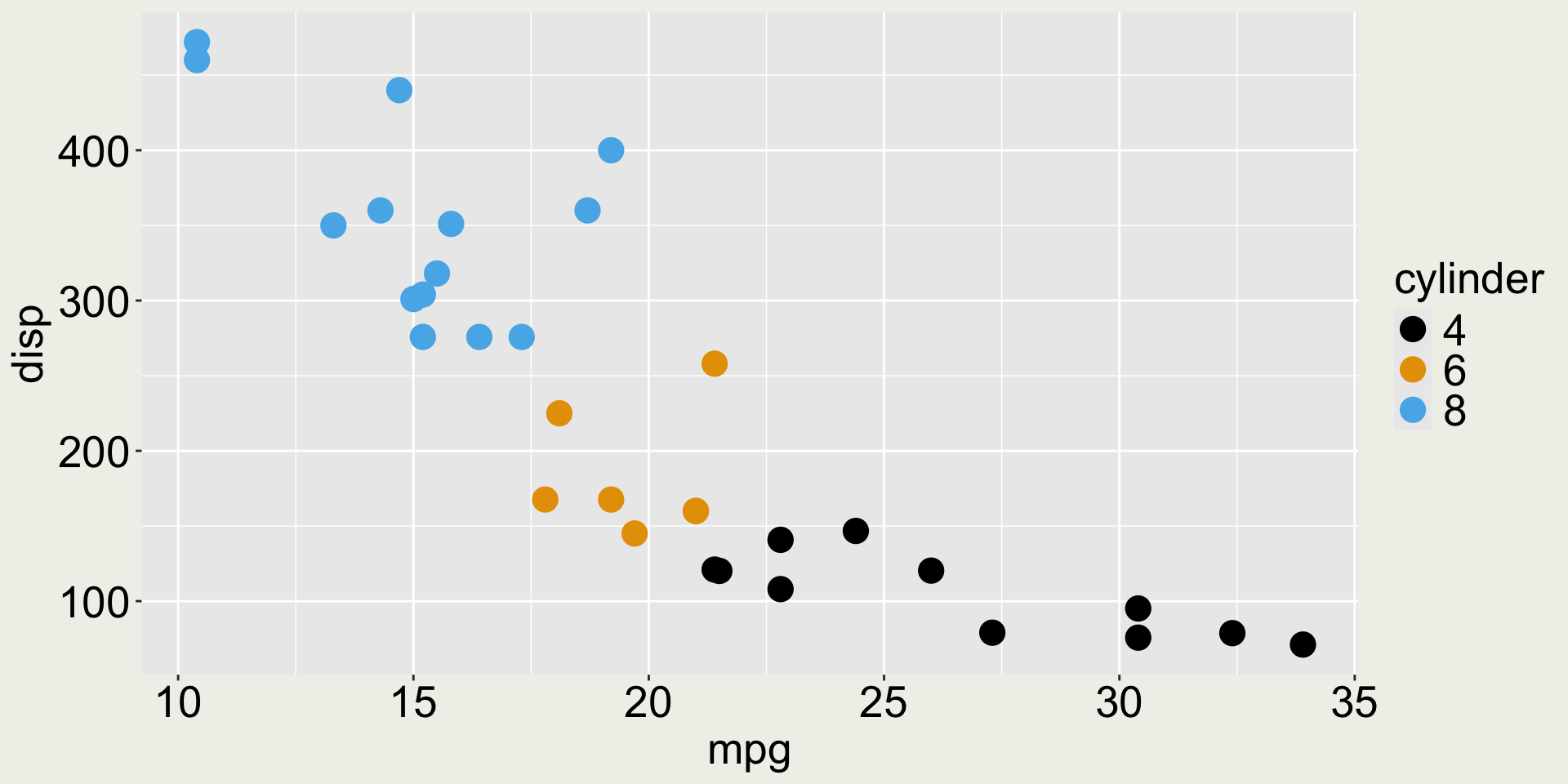
Factor basics
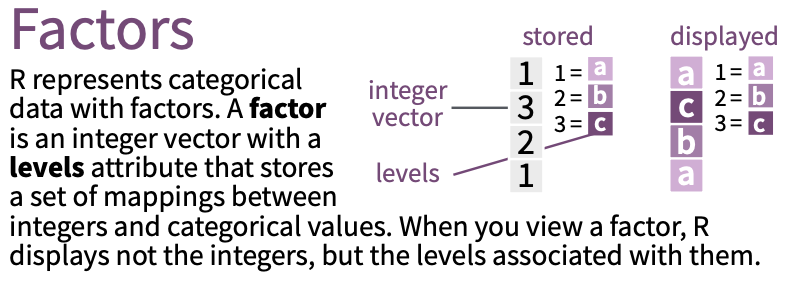
Change a variable to a factor in a tibble
Can you spot the difference before and after?
# A tibble: 32 × 11
mpg cyl disp hp drat wt qsec vs
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 21 6 160 110 3.9 2.62 16.5 0
2 21 6 160 110 3.9 2.88 17.0 0
3 22.8 4 108 93 3.85 2.32 18.6 1
4 21.4 6 258 110 3.08 3.22 19.4 1
5 18.7 8 360 175 3.15 3.44 17.0 0
6 18.1 6 225 105 2.76 3.46 20.2 1
7 14.3 8 360 245 3.21 3.57 15.8 0
8 24.4 4 147. 62 3.69 3.19 20 1
9 22.8 4 141. 95 3.92 3.15 22.9 1
10 19.2 6 168. 123 3.92 3.44 18.3 1
# ℹ 22 more rows
# ℹ 3 more variables: am <dbl>, gear <dbl>,
# carb <dbl># A tibble: 32 × 11
mpg cyl disp hp drat wt qsec vs
<dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 21 6 160 110 3.9 2.62 16.5 0
2 21 6 160 110 3.9 2.88 17.0 0
3 22.8 4 108 93 3.85 2.32 18.6 1
4 21.4 6 258 110 3.08 3.22 19.4 1
5 18.7 8 360 175 3.15 3.44 17.0 0
6 18.1 6 225 105 2.76 3.46 20.2 1
7 14.3 8 360 245 3.21 3.57 15.8 0
8 24.4 4 147. 62 3.69 3.19 20 1
9 22.8 4 141. 95 3.92 3.15 22.9 1
10 19.2 6 168. 123 3.92 3.44 18.3 1
# ℹ 22 more rows
# ℹ 3 more variables: am <dbl>, gear <dbl>,
# carb <dbl><dbl> means double - the variable is a continuous variable <fct> means factor - the variable is discrete/ a factor variable
Factor example 1: change a variable to a factor
The most pedantic way
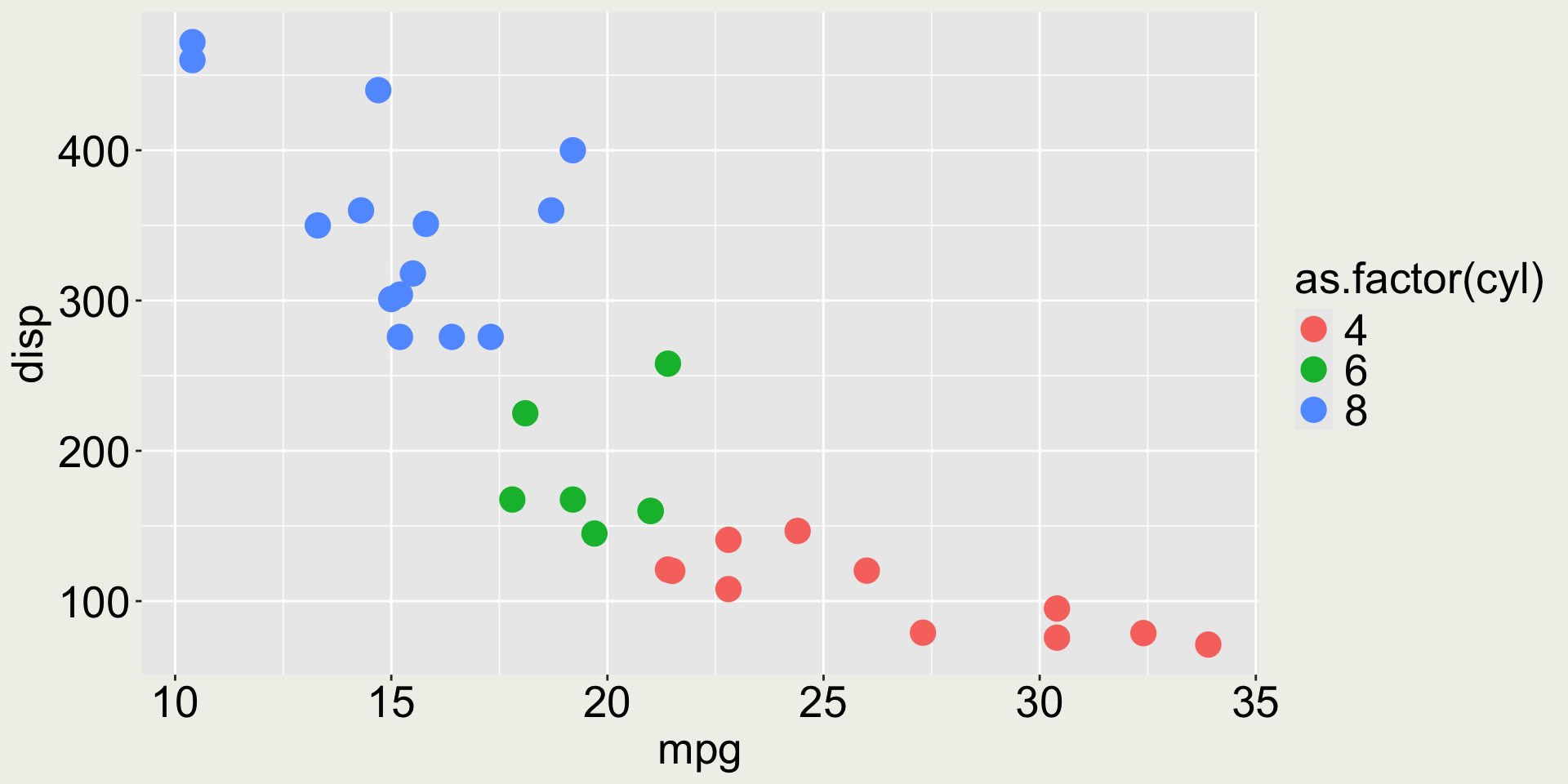
Change the legend name
The legend title is now ugly - we can change it with labs():
Factor example 2: reorder factor levels
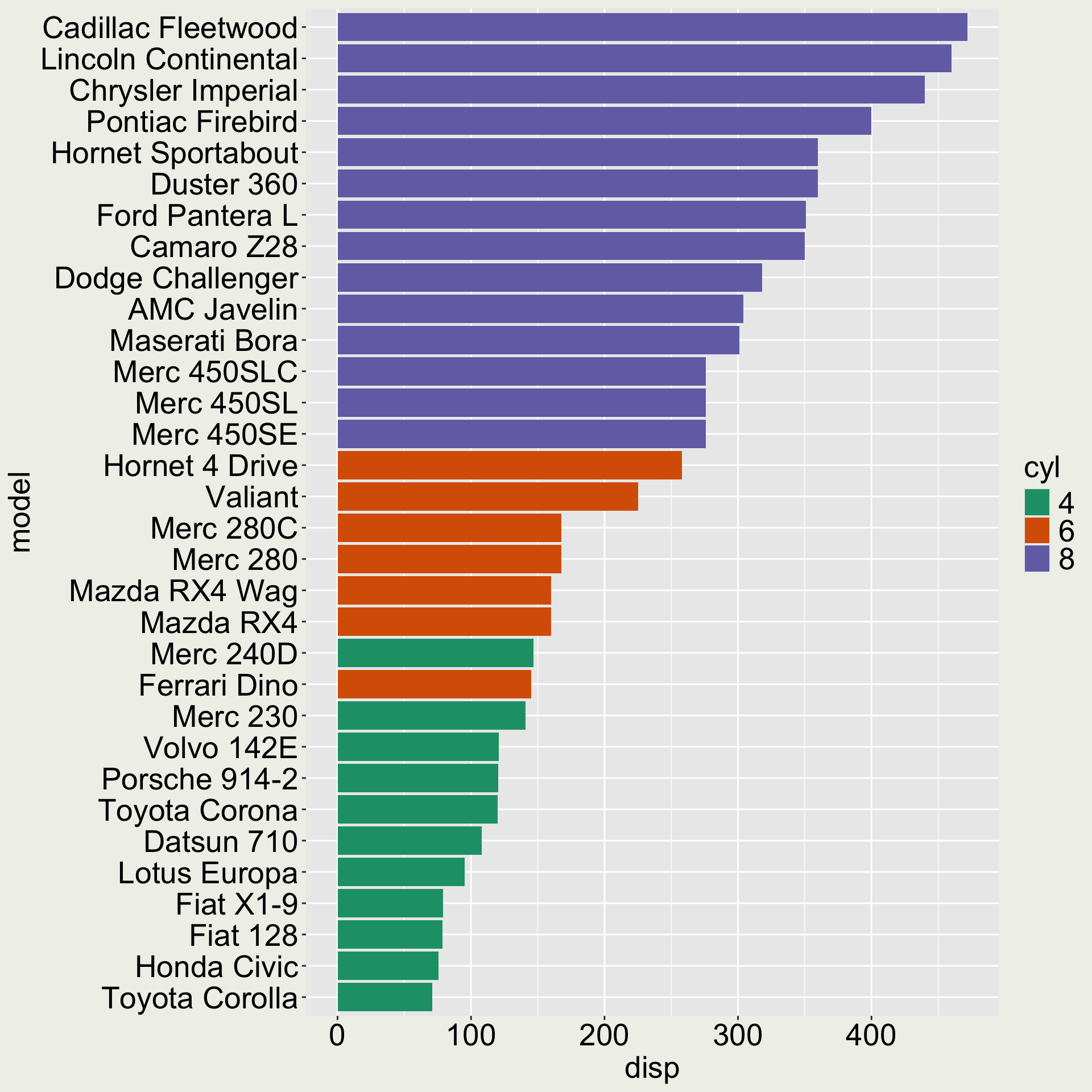
Factor example 2: reorder factor levels
mtcars_tbl <- rownames_to_column(
mtcars, var = "model") |>
as_tibble() |>
mutate(cyl = as.factor(cyl))
mtcars_tbl# A tibble: 32 × 12
model mpg cyl disp hp drat wt qsec
<chr> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Mazd… 21 6 160 110 3.9 2.62 16.5
2 Mazd… 21 6 160 110 3.9 2.88 17.0
3 Dats… 22.8 4 108 93 3.85 2.32 18.6
4 Horn… 21.4 6 258 110 3.08 3.22 19.4
5 Horn… 18.7 8 360 175 3.15 3.44 17.0
6 Vali… 18.1 6 225 105 2.76 3.46 20.2
7 Dust… 14.3 8 360 245 3.21 3.57 15.8
8 Merc… 24.4 4 147. 62 3.69 3.19 20
9 Merc… 22.8 4 141. 95 3.92 3.15 22.9
10 Merc… 19.2 6 168. 123 3.92 3.44 18.3
# ℹ 22 more rows
# ℹ 4 more variables: vs <dbl>, am <dbl>,
# gear <dbl>, carb <dbl>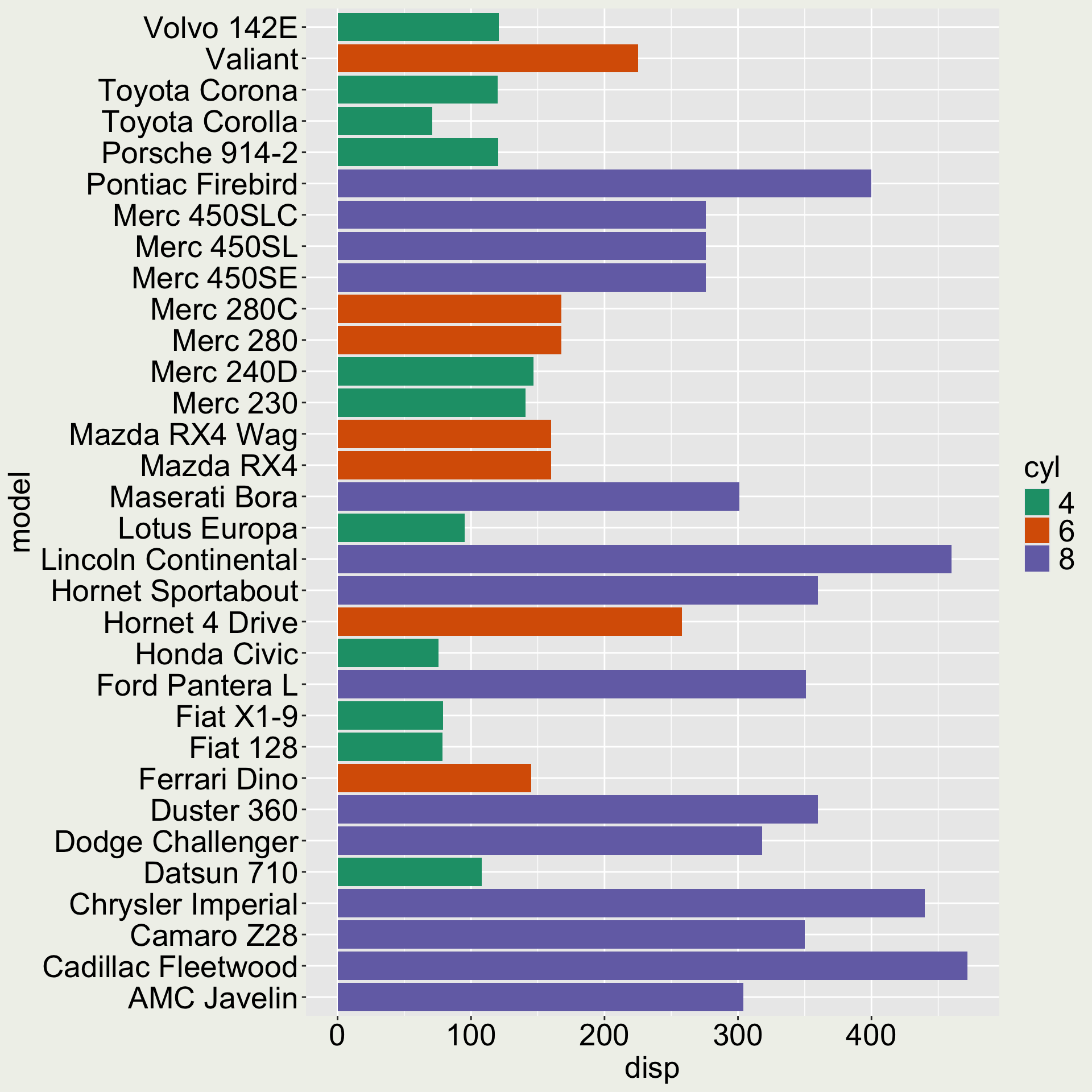
We would like the variable model to be ordered according to disp.
Factor example 2: reorder factor levels
# A tibble: 3 × 2
group values
<chr> <dbl>
1 A 3
2 B 1
3 C 2Two main arguments:
.f: the factor variable you want to reorderx: the variable you want to order by
# A tibble: 3 × 2
group values
<fct> <dbl>
1 A 3
2 B 1
3 C 2Now we see <fct> instead of <chr> for group.
By default .desc = FALSE
- not in descending order, in ascending order: B - C - A (from small to large)
Factor example 2: reorder factor levels

Argument .fun = median means we order by the median of values for each group.
The medians are A: 4, B: 6, C: 5
The order (from small to large) is A - C - B.
[1] A A A B B B C C C
Levels: A C BPreviously we only have one value for each group, so the order is the same as the value itself.
Factor example 2: reorder factor levels

We can change this .fun argument to make it order by min, max, mean, or others:
e.g. Let’s order by the minimum of the each group
The minimum are A: 3, B: 1, C: 2.
The order (from small to large) is B - C - A.
Factor example 2: reorder factor levels
Use fct_reorder() to reorder a factor by another variable
# A tibble: 3 × 12
model mpg cyl disp hp drat wt qsec
<chr> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Mazda… 21 6 160 110 3.9 2.62 16.5
2 Mazda… 21 6 160 110 3.9 2.88 17.0
3 Datsu… 22.8 4 108 93 3.85 2.32 18.6
# ℹ 4 more variables: vs <dbl>, am <dbl>,
# gear <dbl>, carb <dbl>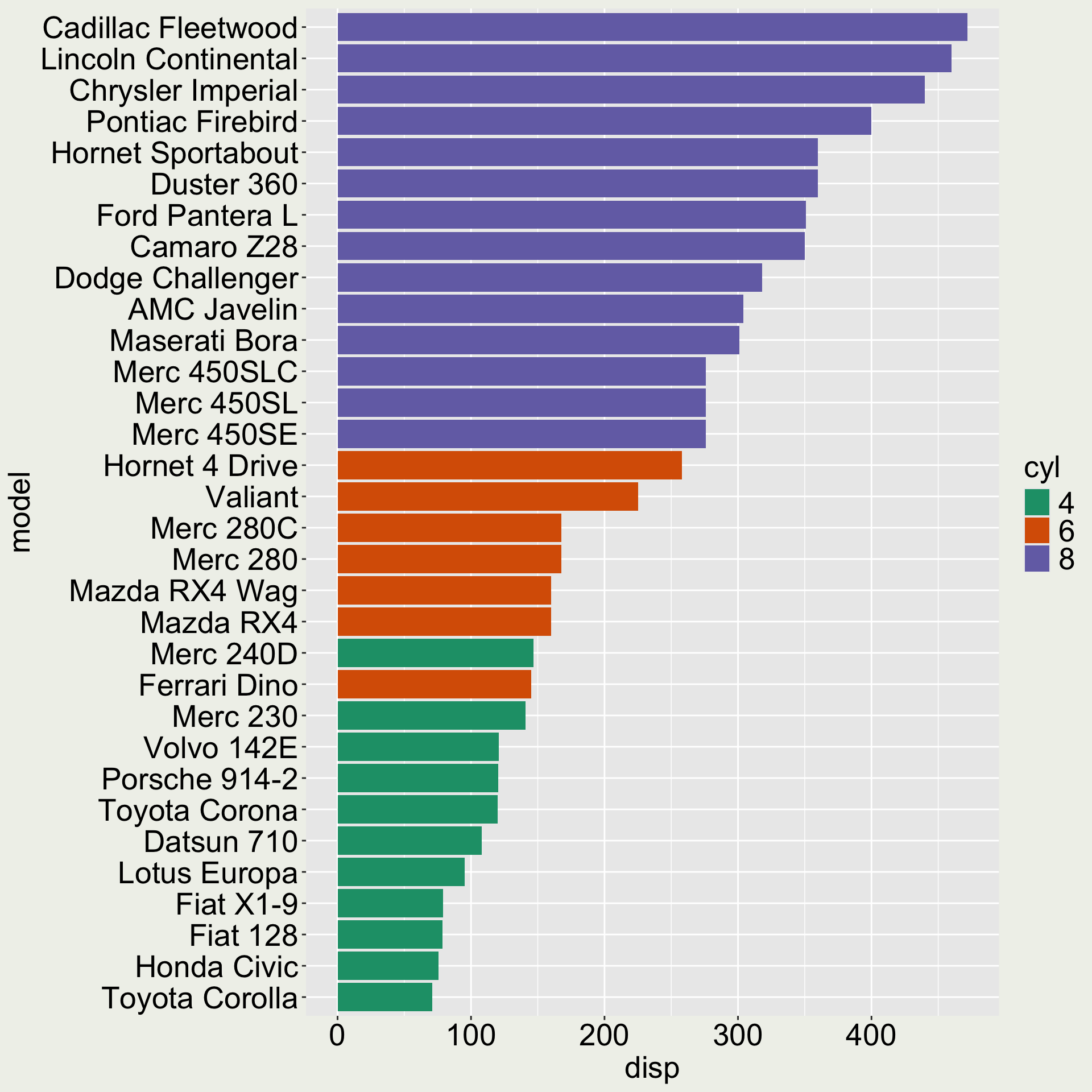
There is also a fct_reorder2() that allows you to order a factor by two variables
Your time
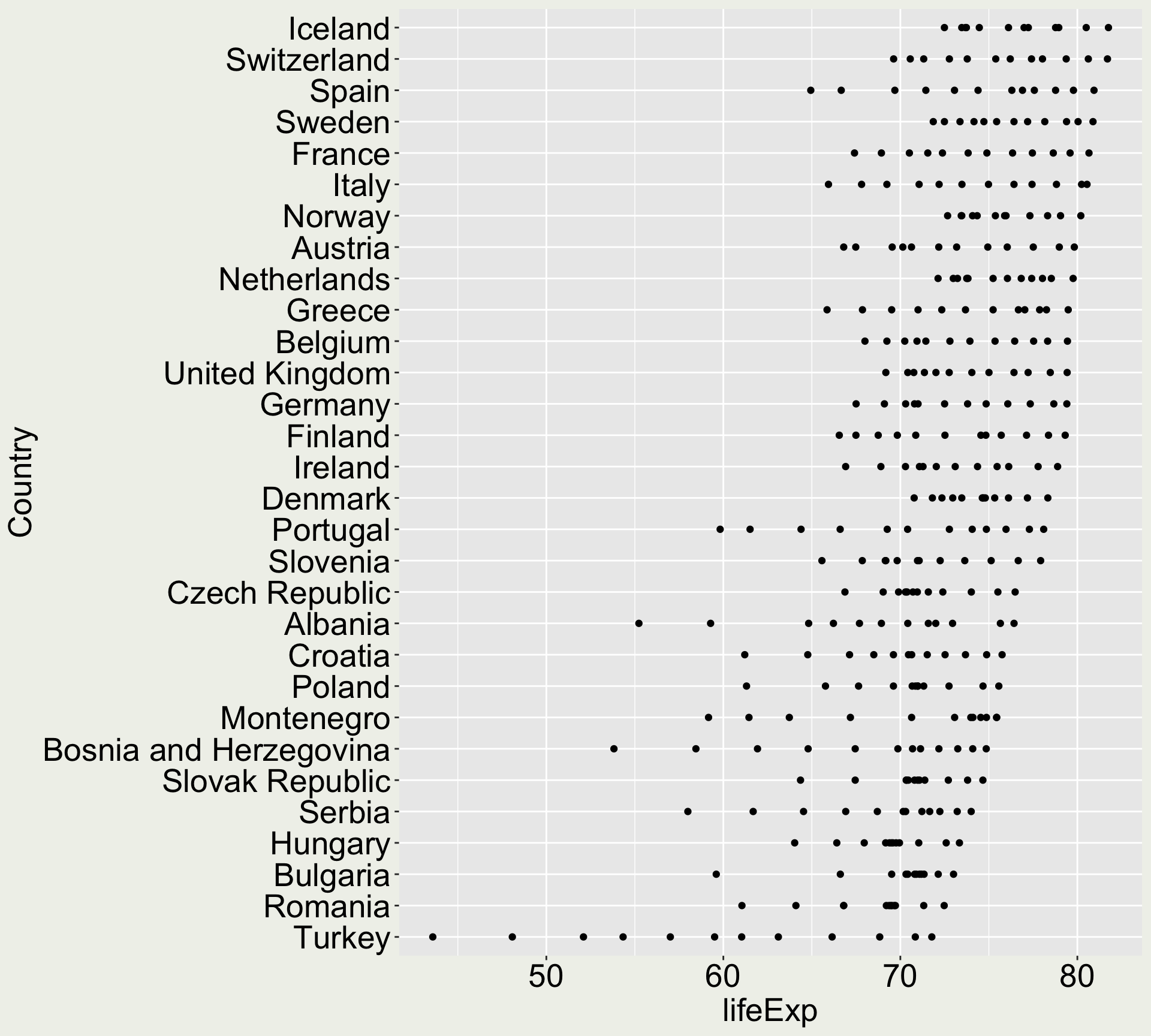
Reproduce this plot:
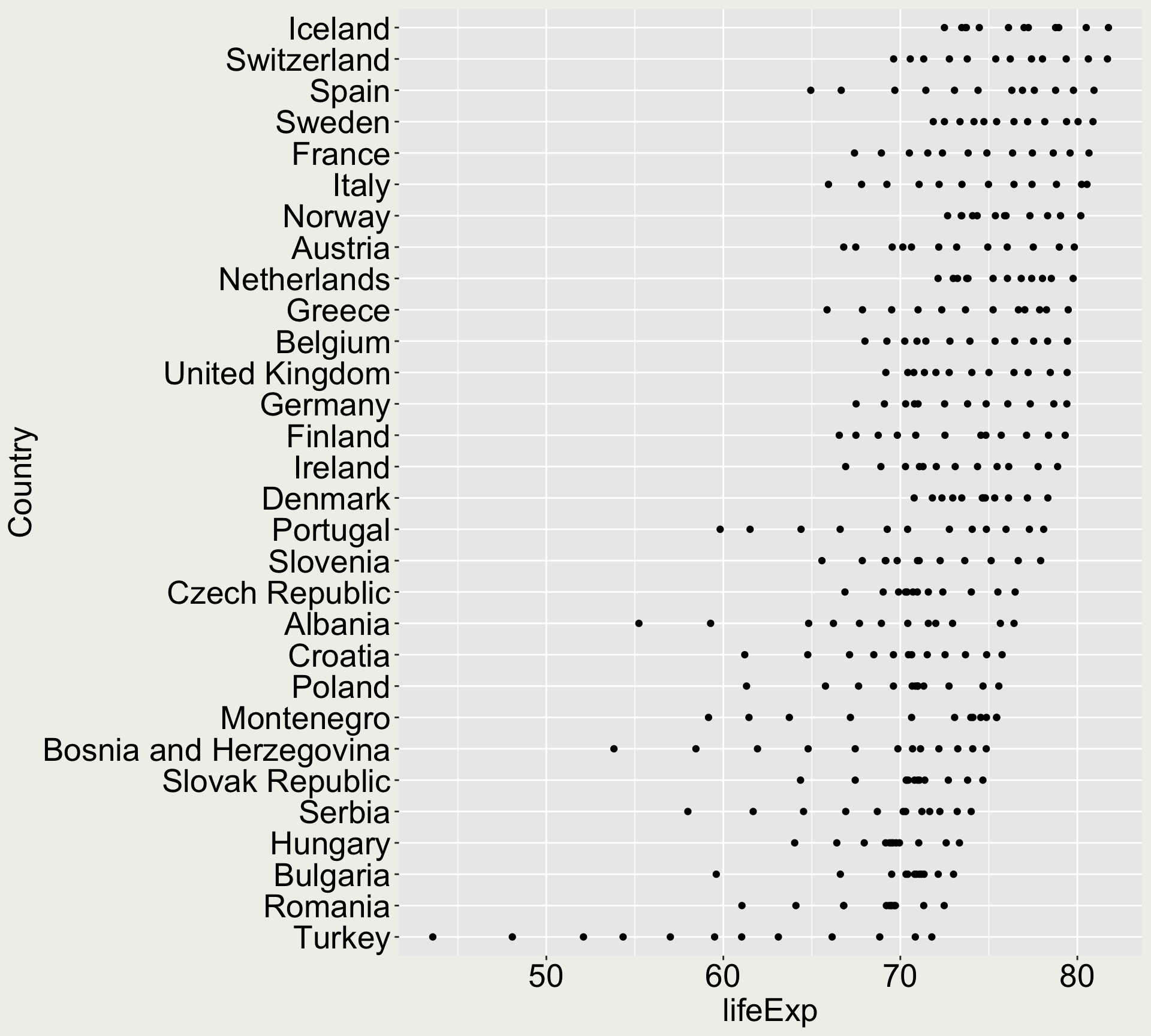
x-axis:
countryy-axis:
lifeExpgeometry: points
We are only using the data in Europe
- How do you subset the data to only European ones?
We order the country by the maximum life expectancy
- How do you do that?
You can play around with order by mean, median, min, etc if you have extra time.
Solution
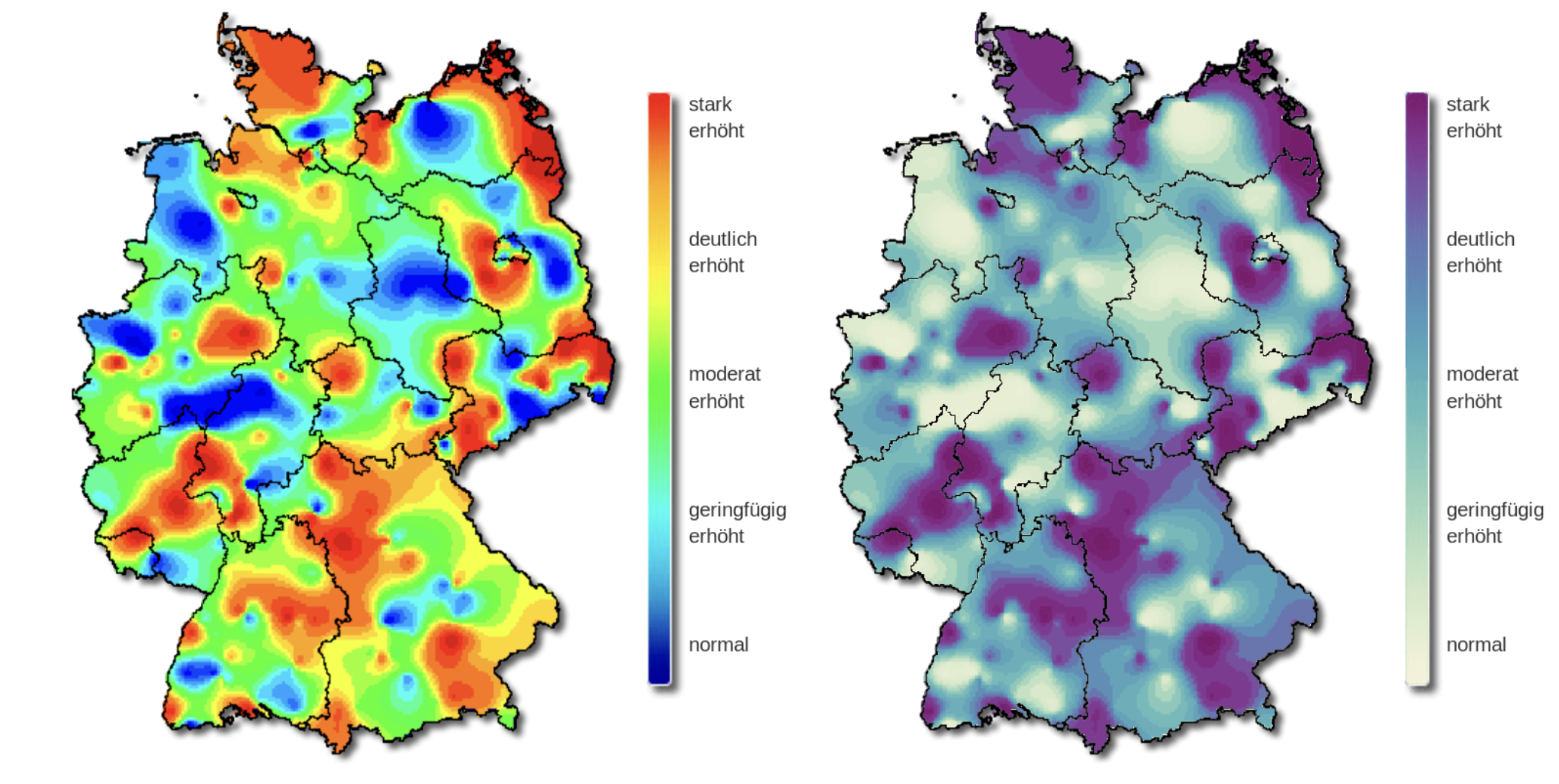
The viridis color palette
Why is viridis a good color palette?
They are perceptually uniform: meaning that values close to each other have similar-appearing colors and values far away from each other have more different-appearing colors, consistently across the range of values.
They are robust to colorblindness, so that the above properties hold true for people with common forms of colorblindness, as well as in grey scale printing.

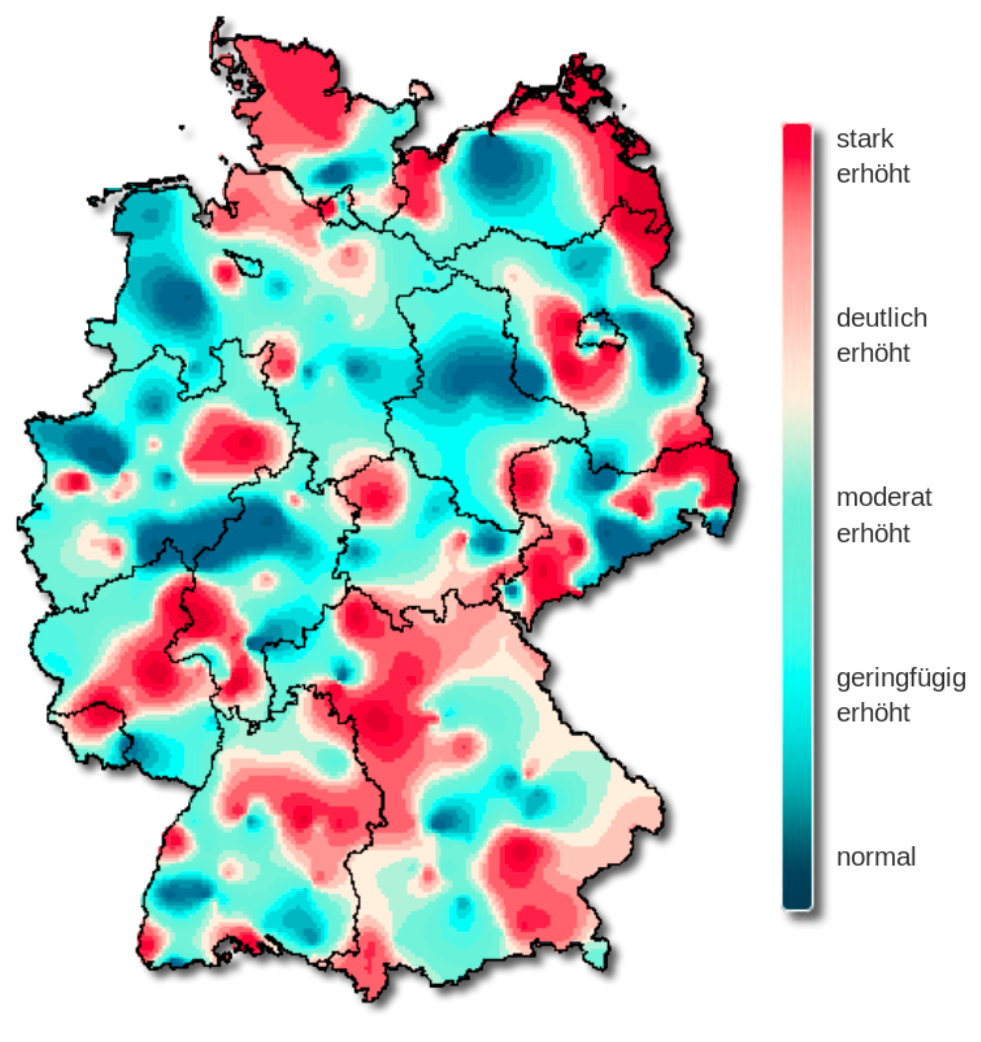
What about the rainbow palette?
The rainbow palette is not uniformly perceived.
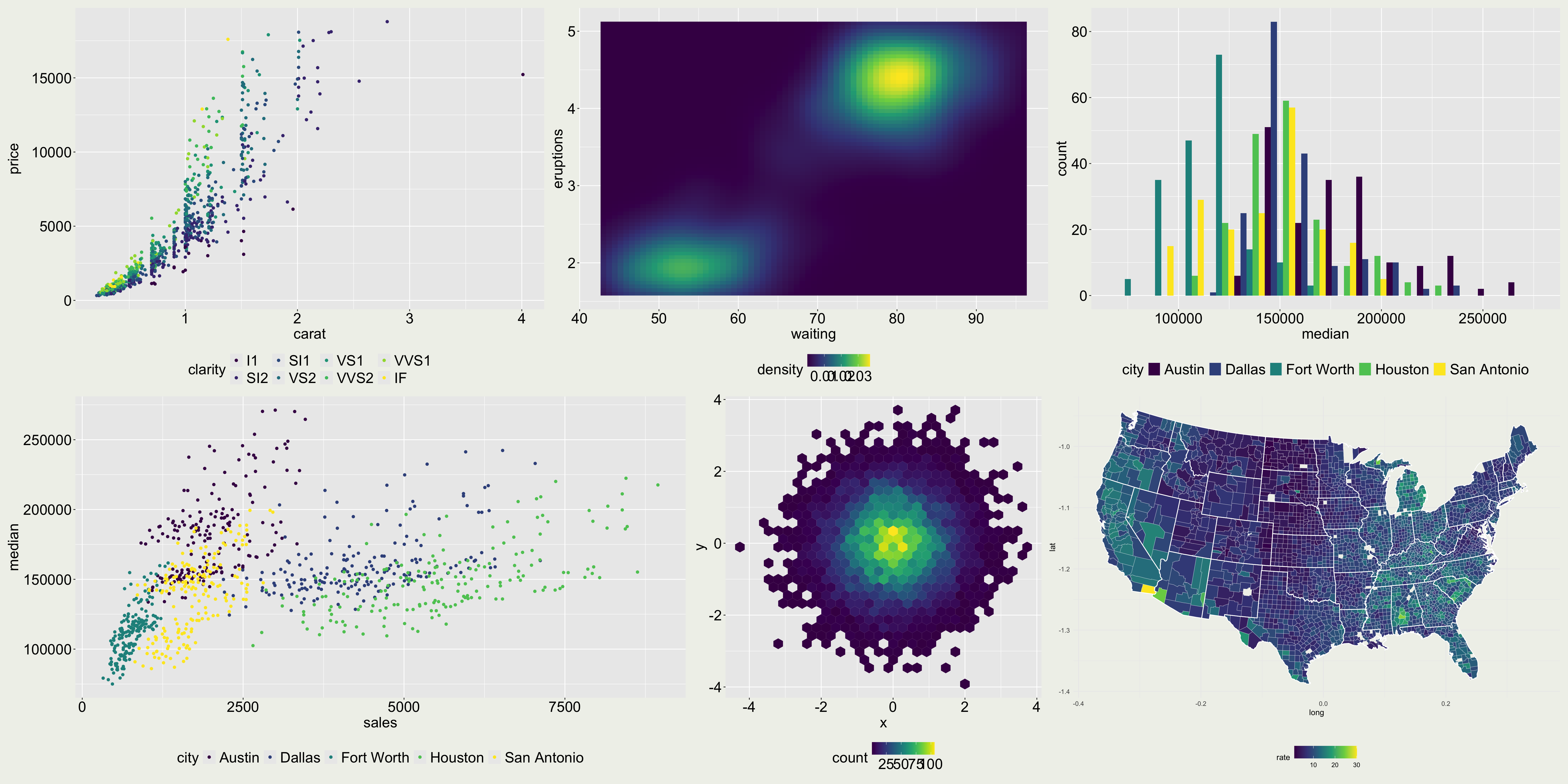
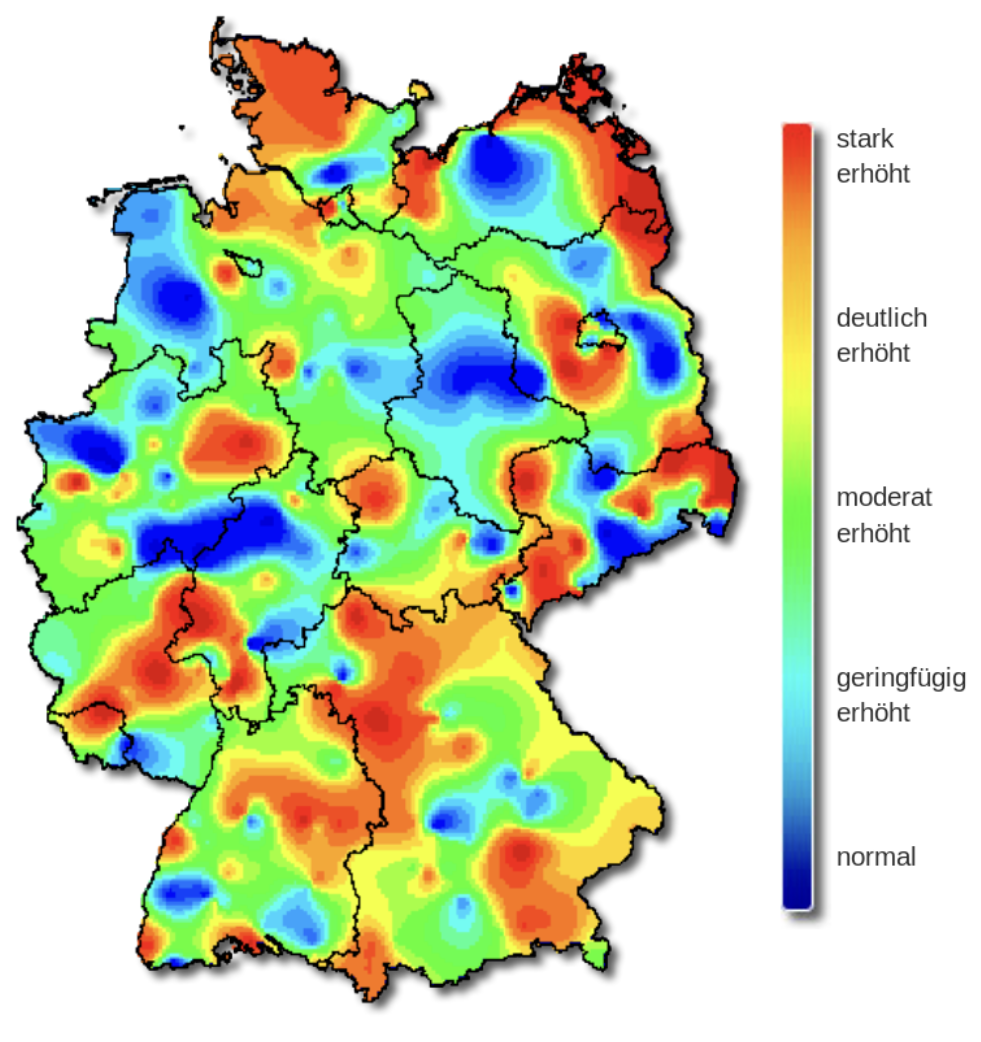
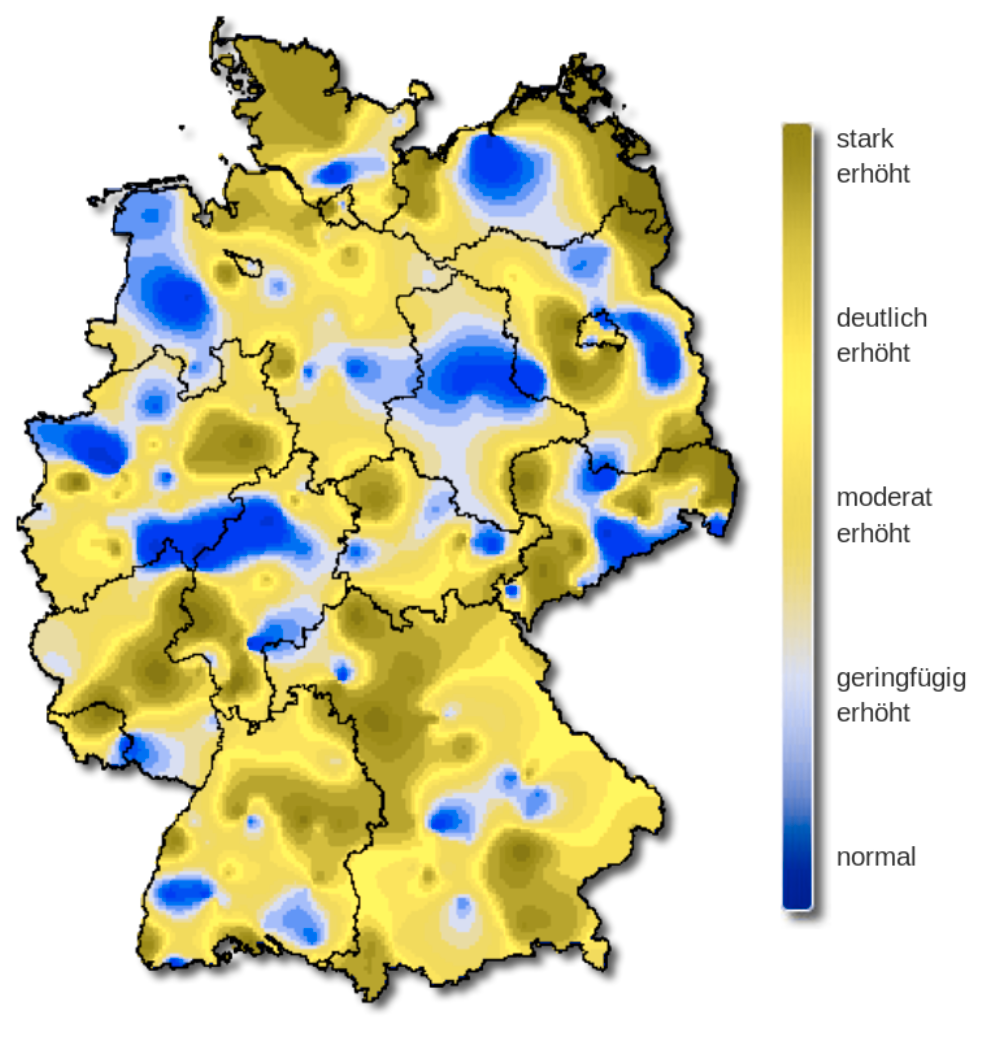
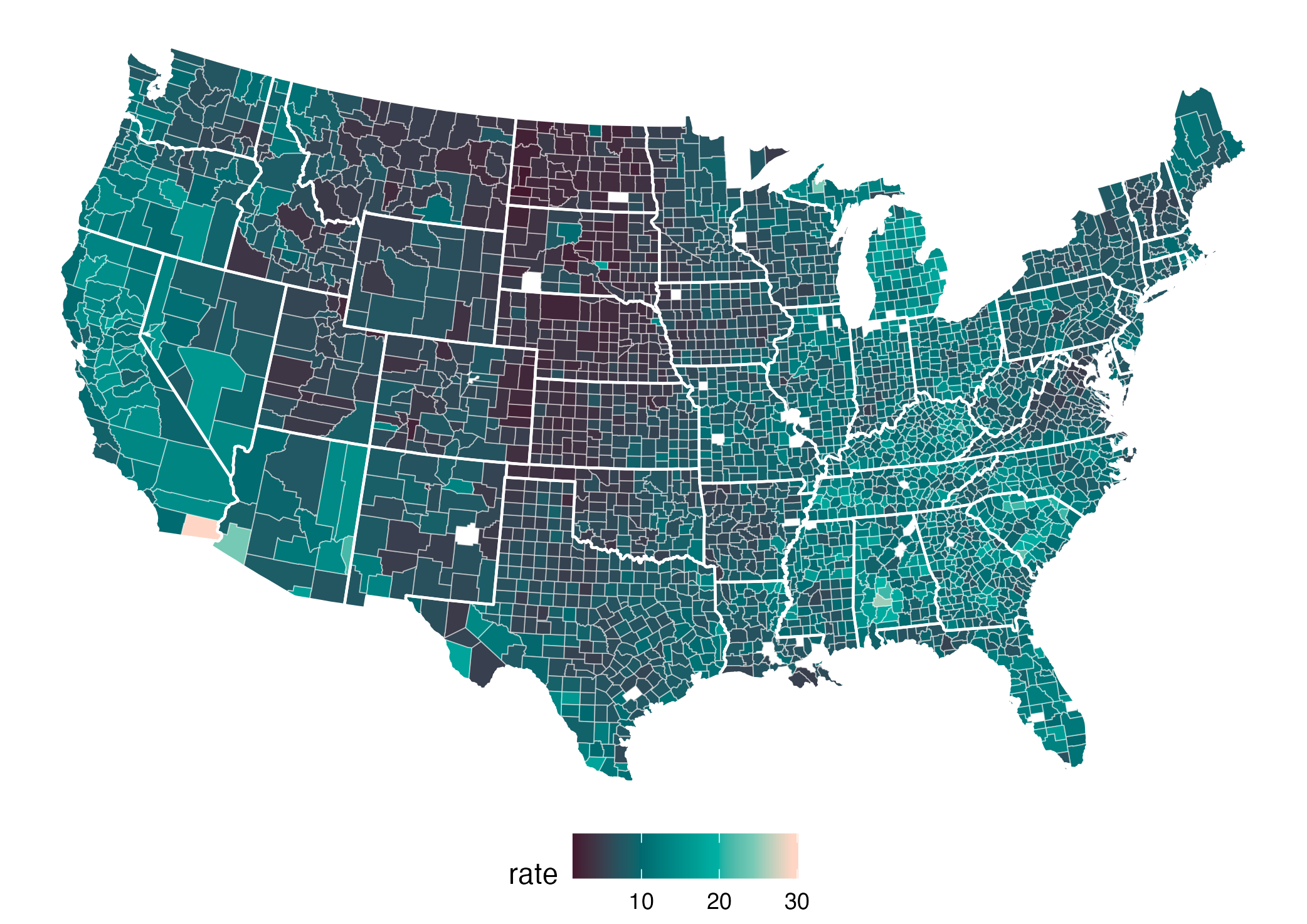
The severity of influenza in Germany in week 8, 2019.
The original color palette (left) is the classic rainbow ranging from “normal” (blue) to “strongly increased” (red).
Color blindness
normal

tritanopia: reduced sensitivity to blue light (extremely rare)

protanopia: reduced sensitivity to red light

deuteranopia: reduced sensitivity to green light (most common)

What does that mean to a color palette?
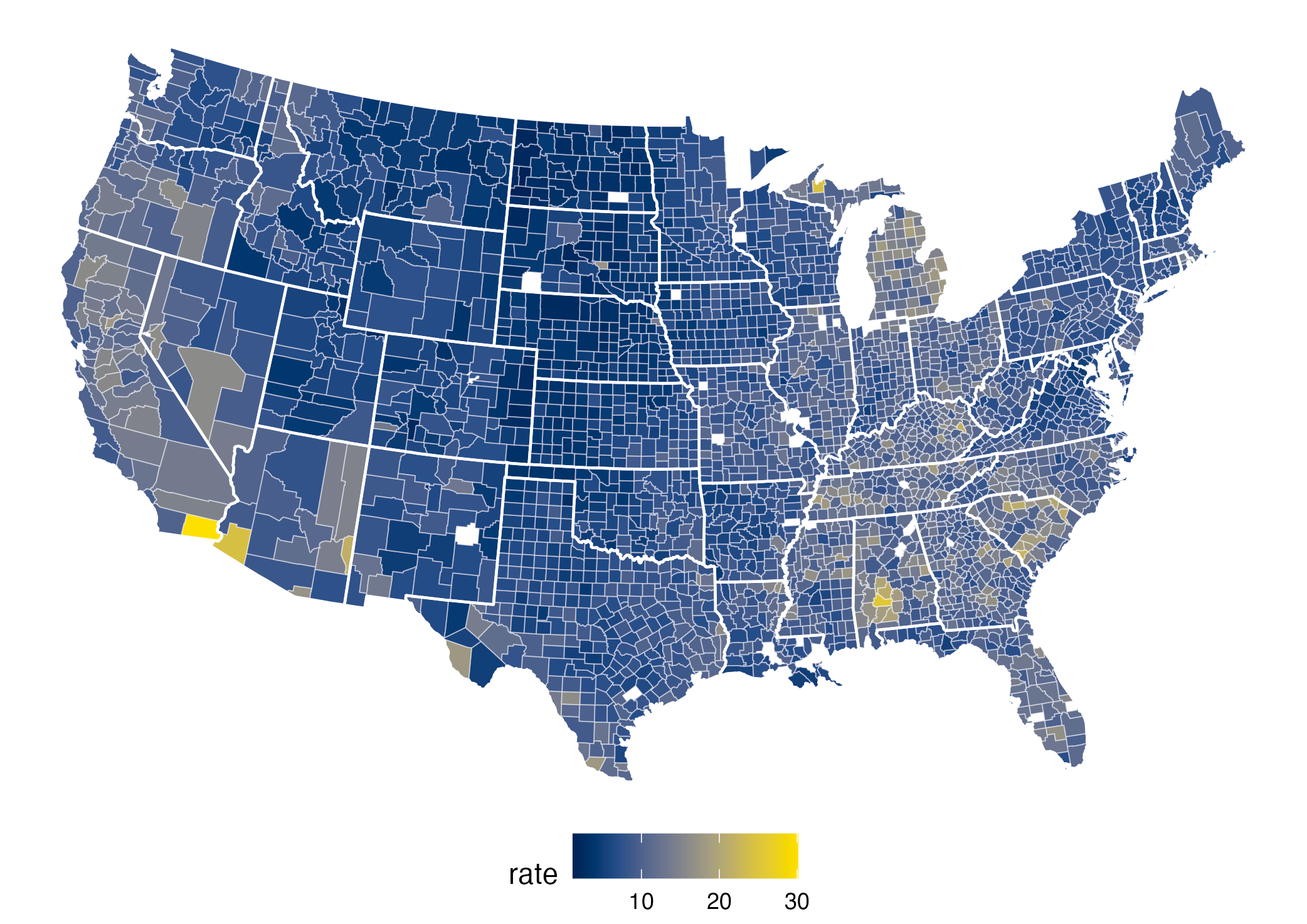
The rainbow palette

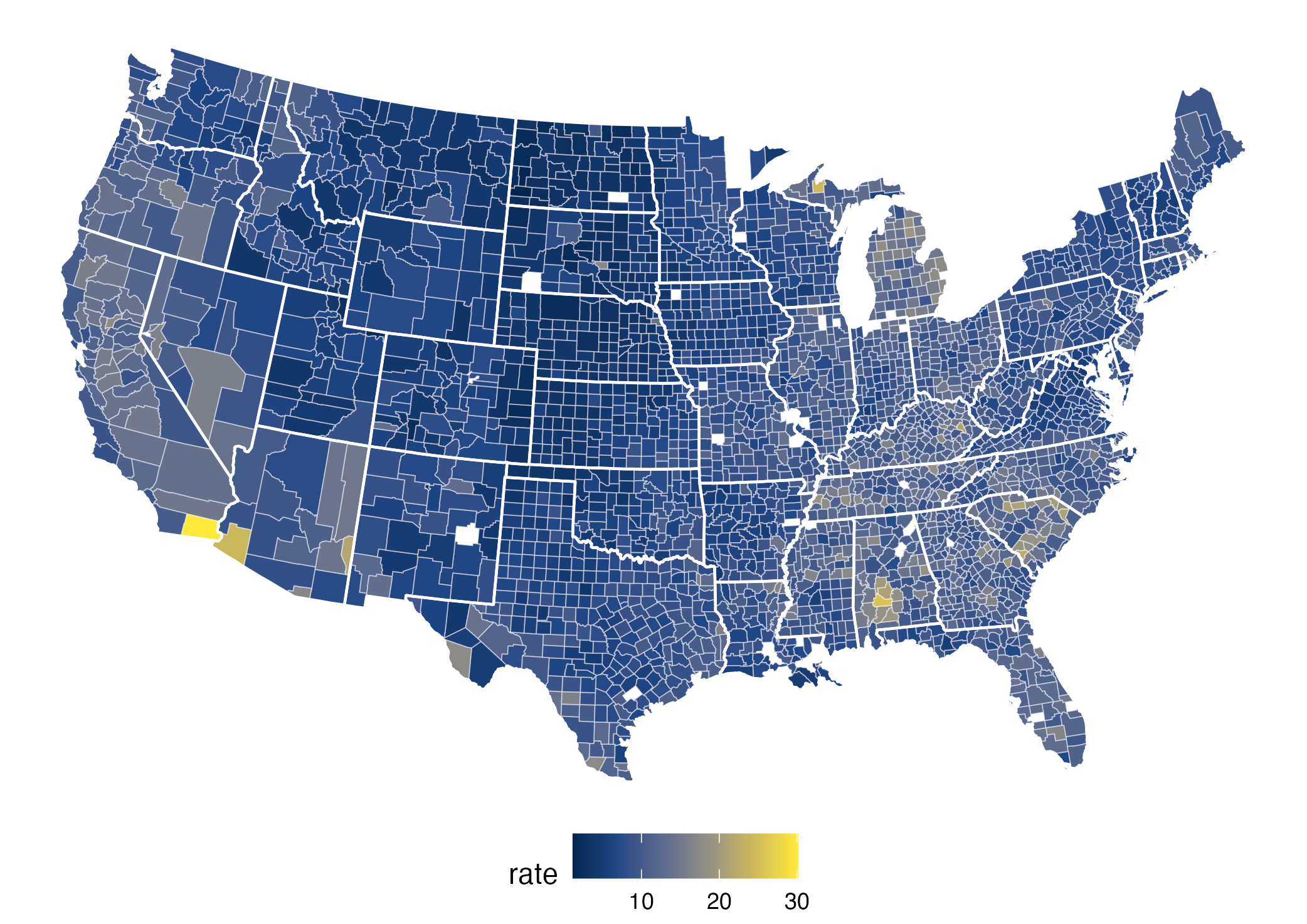
The viridis palette
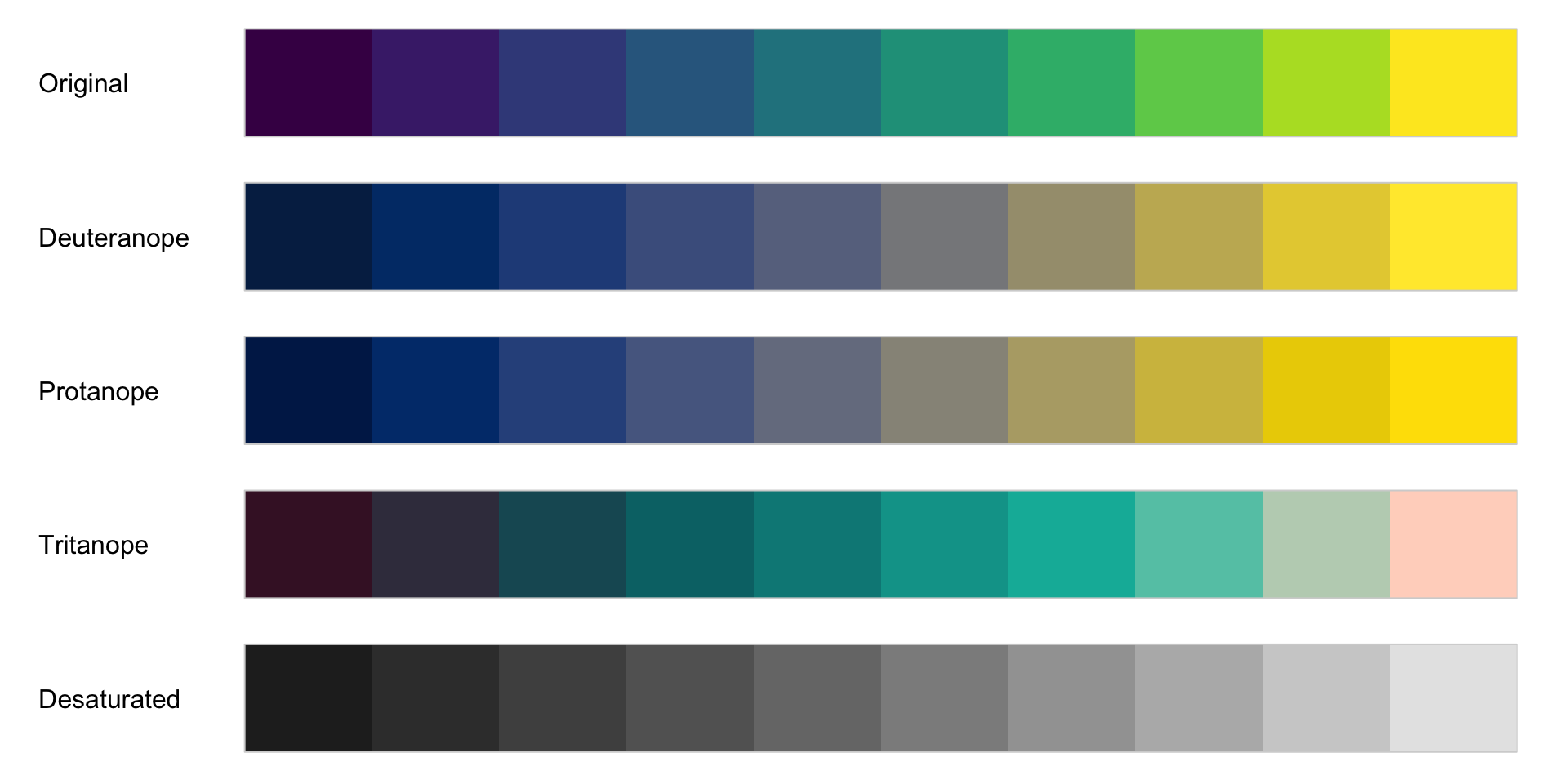
Color blindness affects about 8% of all males and 0.5% of all females!
What does that mean on the plot?
Normal
Deuteranopia
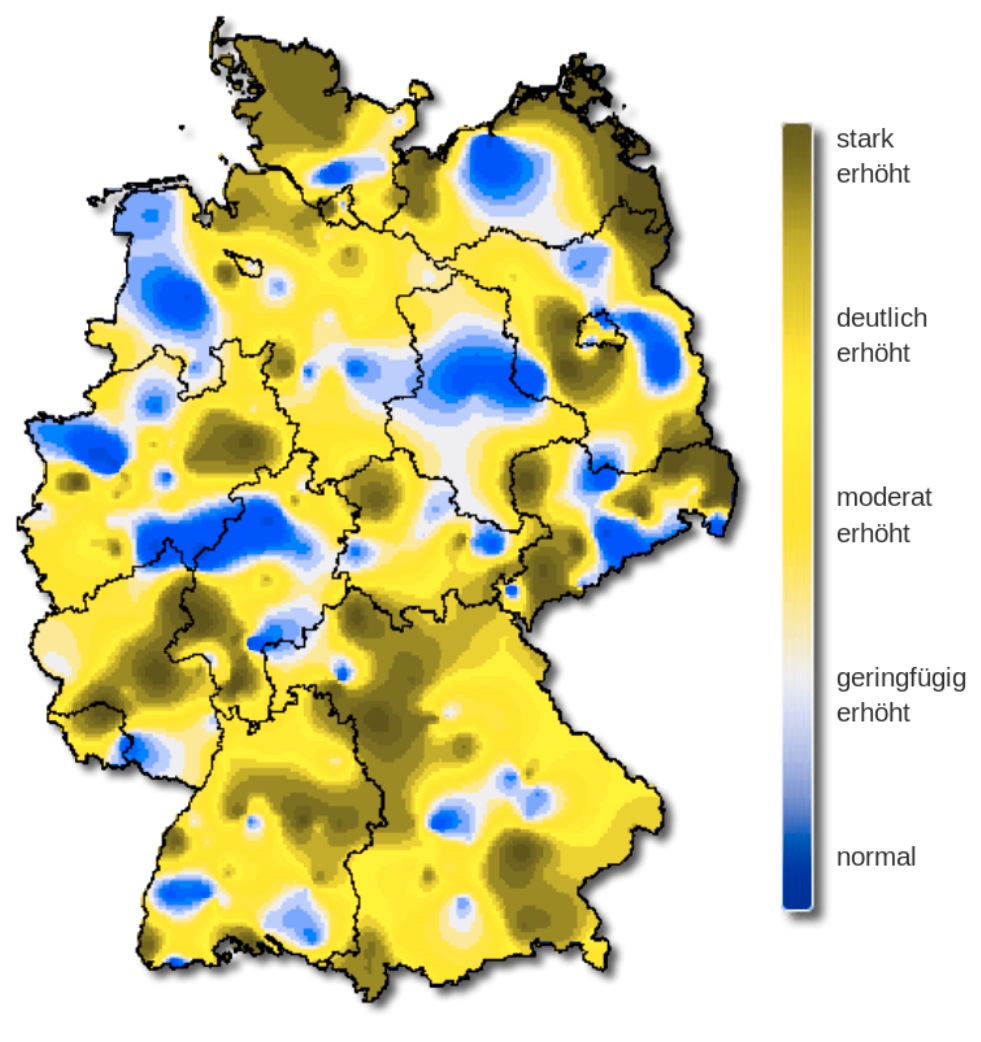
Protanopia
Tritanopia
The rainbow color palette is also not color blind-friendly because baseline color (blue) gets emphasized with deuteranopia and protanopia.
What does that mean on the plot?
normal
tritanopia
protanopia
deuteranopia (most common)
Good color platettes go beyond viridis
Three types of color schemes designed for different types of data:
Qualitative: for categorical information, i.e., where no particular ordering of categories is available and every color should receive the same perceptual weight.
Sequential: for ordered/numeric information, i.e., going from high to low (or vice versa).
Diverging: for ordered/numeric information around a central neutral value, i.e., where colors diverge from neutral to two extremes.

Qualitative color palette: Dark2
Qualitative color palette: Okabe-Ito
New package: ggthemes has many cute themes, scales, and geometries that worth checking out.

To use ggthemes, you need to install it first using install.packages("ggthemes") in the console, and then load it using library(ggthemes) in the script.
Artwork by @allison_horst
Where goes wrong?
Error in p1/p2 : non-numeric argument to binary operator
Error in p1 | p2 : operations are possible only for numeric, logical or complex types
Where goes wrong?
Solution: include library(patchwork) in your script
Easy answer: You forgot to load the patchwork package.
Longer answer:
- By default, the symbol,
/, is an operator for division andpatchworkredefines the symbol to combine the two plots up-and-down.
- When the package is not loaded, what R thinks is that
p1andp2are not numbers that I can do arithmetic, so let me stop and throw an error message.
Example: Patchwork
Example: Patchwork
Your time
This is a plot I show you in week1 hello-world.pdf. Can you use the mtcars data with things you’ve learnt from ggplot2 to create the exact same plot?
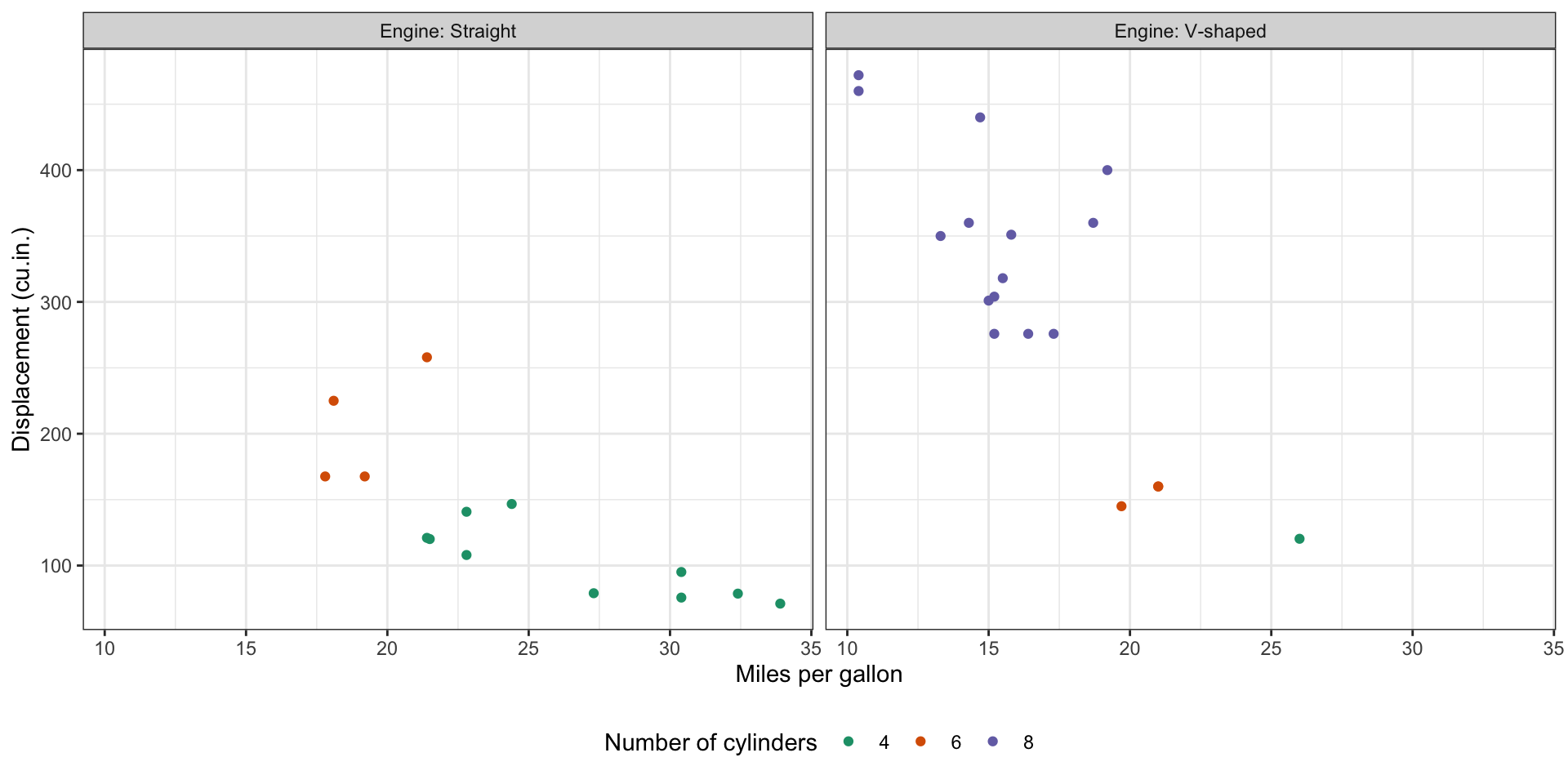
There are some hints in the next slides to guide you make this plot step-by-step.
Your time
Base plot: Start with a base plot that map the variables in
mtcarsto the x, y-axis, color, and facet.Color: The color seems to be mapped to a continuous value. Is it the best choice? How would you change it? What’s the
scale_xxx_xxx()function to change to a different color palette.Facet: The facet header (0 and 1) are not informative, how would you change it. Maybe we can recode 0 and 1 to its actual meaning. How would you do that?
Labels: Use a more informative x and y axis title, and legend name
Theme: Play around with theme and arrange the legend position to bottom